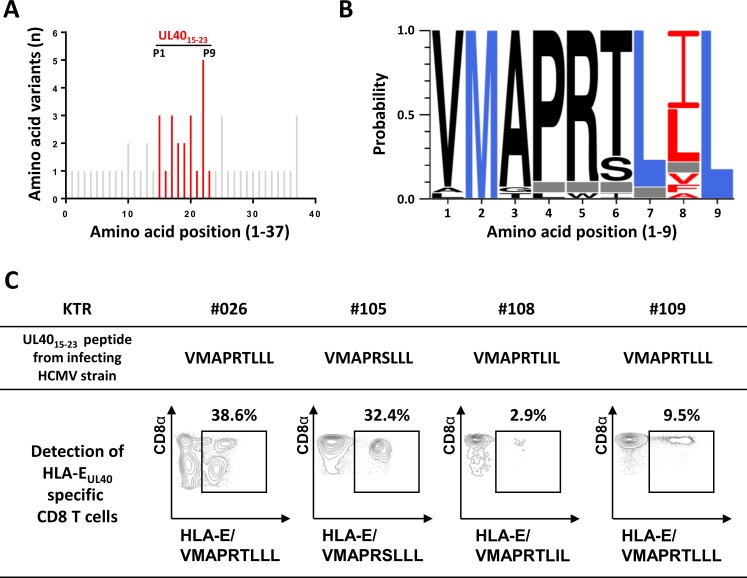
Fig 3. HCMV strain-dependent variability of UL4015-23 sequences and HCMV strain-specific HLA-EUL40 T-cell response in hosts.
(A, B) Genomic DNAs isolated from HCMV positive blood samples in HCMV+ transplant recipients (n = 25) were sequenced for the identification of UL40 protein (amino acids 1–221) provided by the circulating HCMV strains. (A) Amino acid variability, expressed as a number of amino acid variants, within the HLA-E-binding peptide (UL4015-23, shown in red) among the sequence for HCMV UL40 signal peptide (UL401-37, shown in grey). A total of 32 UL40 sequences from 25 hosts were analysed. UL40 protein sequence from the Merlin HCMV clinical strain was used as reference. Positions 1 to 9 of residues in the HLA-E-binding peptide (UL4015-23) are indicated. (B) Sequence LOGO of the UL4015-23 HLA-E-binding peptide from 25 transplanted hosts. The height of the letter is proportional to the frequency of each amino acid in a given position (P1 to P9). Major anchor residues for binding in the HLA-E peptide groove are indicated in blue. Red letters highlight the important variability observed in position 8 of the HLA-E-binding peptide. Grey boxes correspond to a constitutive deletion of the corresponding amino acid in the UL40 sequence from the infecting viral strain. (C) Representative dot plot analyses showing the detection of strain-specific anti-UL40 HLA-E-restricted CD8 T-cell responses in 4 KTRs (KTR#026, #105, #108 and #109). Frequencies (%) of the HLA-EUL40-specific T cells among total circulating αβ CD8 T cells are indicated.