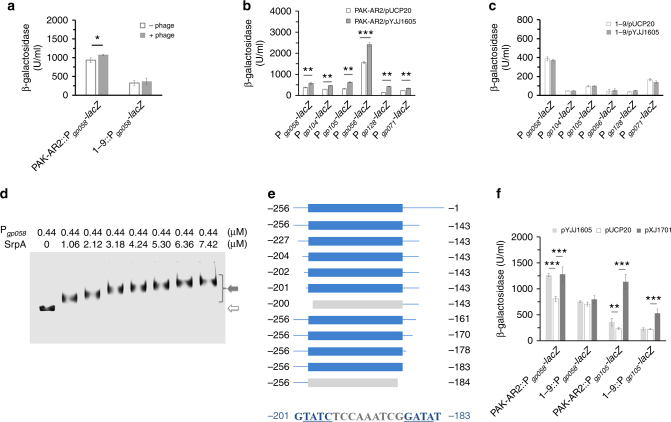
Fig. 3.
Regulatory function of the SrpA. a Phage K5 infection improves the transcription of the phage RNA polymerase gene gp058 in PAK-AR2. b Transcription profiles of the indicated phage genes in PAK-AR2 carrying pYJJ1605, which expresses the phage RNA polymerase gene gp058 driven by its native promoter. c Transcription profiles of the indicated phage genes in 1–9 carrying pYJJ1605. d EMSA assay for the SrpA binding of the gp058 promoter. The gray arrow indicates bound DNA fragments. The blank arrow indicates free DNA fragments. e Mapping of the SrpA-binding sequence by EMSA assay. Fragments with the intact binding site (blue color) showed positive bindings. Fragments with the defective binding site (gray color) showed no bindings. Arabic numbers represent the distances (bp) upstream from the start codon ATG of the srpA gene. f β-galactosidase activity assay of strains PAK-AR2 and 1–9 harboring either the Pgp058-lacZ (with a SrpA-binding site) or Pgp105-lacZ (without a SrpA-binding site) reporter fusion. The plasmid pYJJ1605 and pXJ1701 express the phage RNA polymerase gene gp058 driven by its native promoter and the Plac promoter, respectively. The experiments were independently replicated three times and one-way ANOVA was used to examine the mean differences between the data groups (a–c, f). *P < 0.05. **P < 0.01. ***P < 0.001. Error bars show standard deviations