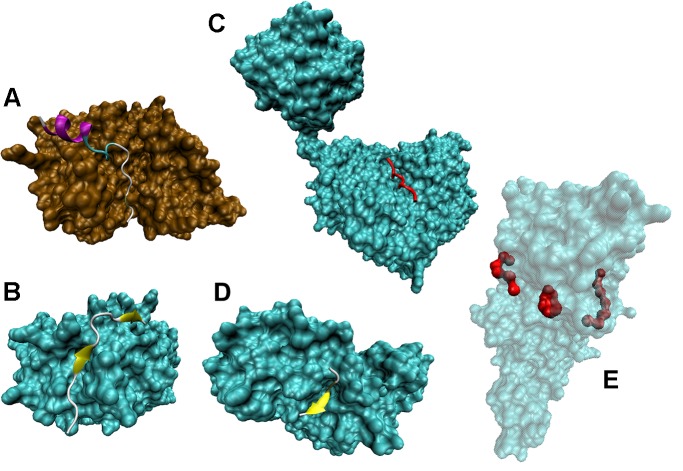
FIGURE 4.
Disordered nature of the PTM sites in target proteins. (A) Crystal structure of a complex between a 20-amino acid peptide derived from the heat stable protein kinase inhibitor (PKIα) and the catalytic subunit of cyclic AMP-dependent protein kinase (cAPK) (PDB ID: 2CPK) (Knighton et al., 1991). (B) Crystal structure of a complex between a 23 amino acid cell-permeable peptide and a protein Ser/Thr phosphatase-1 (PDB ID: 4G9J) (Chatterjee J. et al., 2012). (C) Crystal structure of a complex between polypeptide N-acetylgalactosaminyltransferase 2 and a 13 amino acid peptide EA2 (PDB ID: 2FFU) (Abraham and Podell, 1981). (D) Crystal structure of a complex between human mitochondrial NAD-dependent deacetylase sirtuin-3 and a 12 amino acid peptide derived from mitochondrial acetyl-coenzyme A synthetase 2-like (PDB ID: 3GLT) (Jin et al., 2009). (E) Crystal structure of a complex between a protein arginine N-methyltransferase 1 and a 19 amino acid substrate peptide (PDB ID: 1OR8) (Zhang and Cheng, 2003).