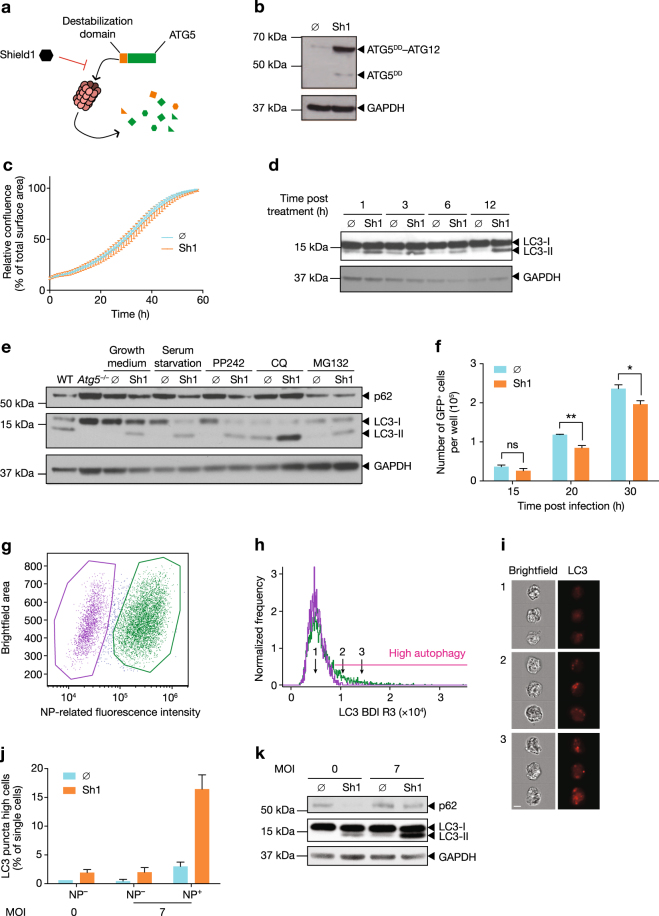
Fig. 1. Stabilization of ATG5 in Atg5–/– cells enables experimental control of autophagy.
a Schematic representation of Shield1 (Sh1) stabilization of ATG5 illustrates the rescue of destabilization domain (DD)-fused ATG5 (ATG5DD). b ATG5DD-expressing Atg5–/– cells were treated with ethanol vehicle (∅) or Sh1 for 20 h, followed by immunoblot analysis with anti-ATG5 antibody. c ATG5DD-expressing Atg5–/– cells were treated with Sh1 and images were obtained every hour for 60 h to assess cell growth. Points depict mean confluence at interval time and error bars depict standard deviation. d ATG5DD-expressing cells were treated for the indicated times with Sh1 or vehicle (∅). Protein extracts were subjected to immunoblot analysis using anti-LC3 and anti-GAPDH antibodies. e In the presence or absence of Sh1, cells were exposed to serum deprivation, an inhibitor of the mammalian target of rapamycin (PP242), a proton pump inhibitor (chloroquine, CQ) or a proteasome inhibitor (MG132). Wild-type (WT) and Atg5–/– MEFs (Atg5–/–) were used as positive and negative controls, respectively. After 4 h of culture, protein extracts were subjected to immunoblot analysis using anti-p62, anti-LC3 and anti-GAPDH antibodies. f ATG5DD cells, pretreated or not with Shield1 (Sh1), were infected with GFP-expressing chikungunya virus at an MOI of 0.1. The number of green cells were monitored through live imaging. Graph shows mean and standard deviation of three biological replicates, and data are representative of three experiments. ns, not significant; *q < 0.01, **q < 0.01 (two-tailed unpaired t-test followed by Holm’s multiple testing correction). g–k ATG5DD expression was stabilized by Sh1 treatment and cells were infected with influenza A virus (IAV). After 20 h, cells were fixed, permeabilized and stained using anti-NP antibody to quantify IAV infection and anti-LC3 antibody to visualize autophagy puncta. Imaging flow cytometry permitted gating based on NP expression (representative dot plot, g) and quantification of autophagic vesicles using the bright detail intensity R3 (BDI R3, histogram, h). i Three representative images of single cells with three different BDI R3, corresponding to the indicated numbered arrows in h, are shown. The topography of LC3 within cells is shown in red. Scale bar, 10 μm. j Graphs plot mean percentage of cells with high autophagic vesicle content and standard deviation; analysis is based on images captured from >10,000 cells per experiment (n = 2 experiments). k Cells were analysed by immunoblot for p62, LC3 and GAPDH expression