Figure 3. Stendomycin inhibits TIM23-dependent translocation in vivo.
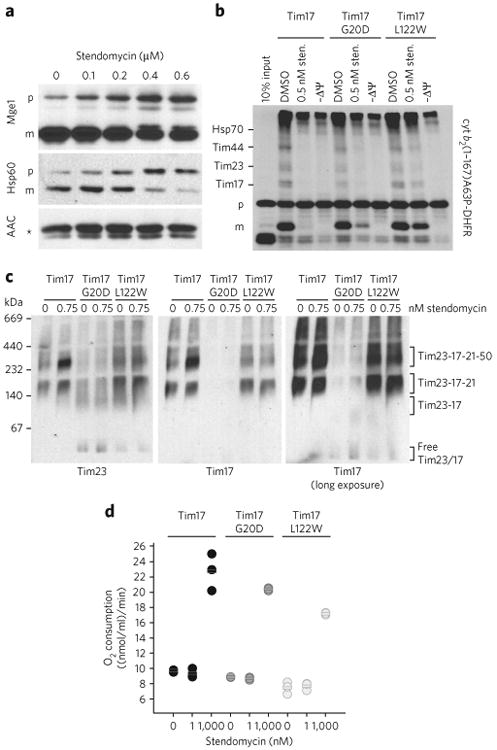
(a) In vivo accumulation of TIM23 complex substrates precursors. Wild-type yeast cells were grown in YPEG media and treated with the indicated concentration of stendomycin for 24 h. The accumulation of Hsp60 and Mge1 precursors was analyzed by immunoblot. The TIM22 complex substrate ACC was used as loading control. p, precursor; m, mature; * marks an unspecific reaction of the antibody. (b) cyt b2(1-167)A63P-DHFR was imported into mitochondria with the addition of 0.5 nM stendomycin or 1% DMSO. The membrane potential was disrupted by CCCP (−ΔΨ). Protein import was arrested by the addition of methotrexate, and cyt b2 (1-167)A63P-DHFR was crosslinked to components of the TIM23 import pathway by DSS. Crosslinked intermediates are indicated. p, precursor; m, mature. Assignment of crosslinked bands was performed as described21,22. (c) Mitochondria isolated from wild-type, Tim17G20D and Tim17L122W yeast strains were treated with 0.75 nM stendomycin for 15 min, solubilized with digitonin and separated by BN–PAGE. The TIM23 complex was analyzed by immunoblot using antibodies targeting Tim23 and Tim17 (see also Supplementary Fig. 2b). (d) Oxygen consumption of isolated mitochondria from indicated yeast strains was measured with an oxygen electrode. Respiration was initiated by the addition of NADH. 1 or 1,000 nM stendomycin or 1% DMSO was added once a stable respiration had been established. Respiration rates were quantitated based on the DMSO control (see Supplementary Fig. 3 for representative oxygraph plots). Bars represent mean values of n = 3 biological replicates (separate mitochondrial preparations), which are shown as circles (black, wild type; dark grey, Tim17G20D; light grey, Tim17L122W. Full gel scans are in Supplementary Information.
