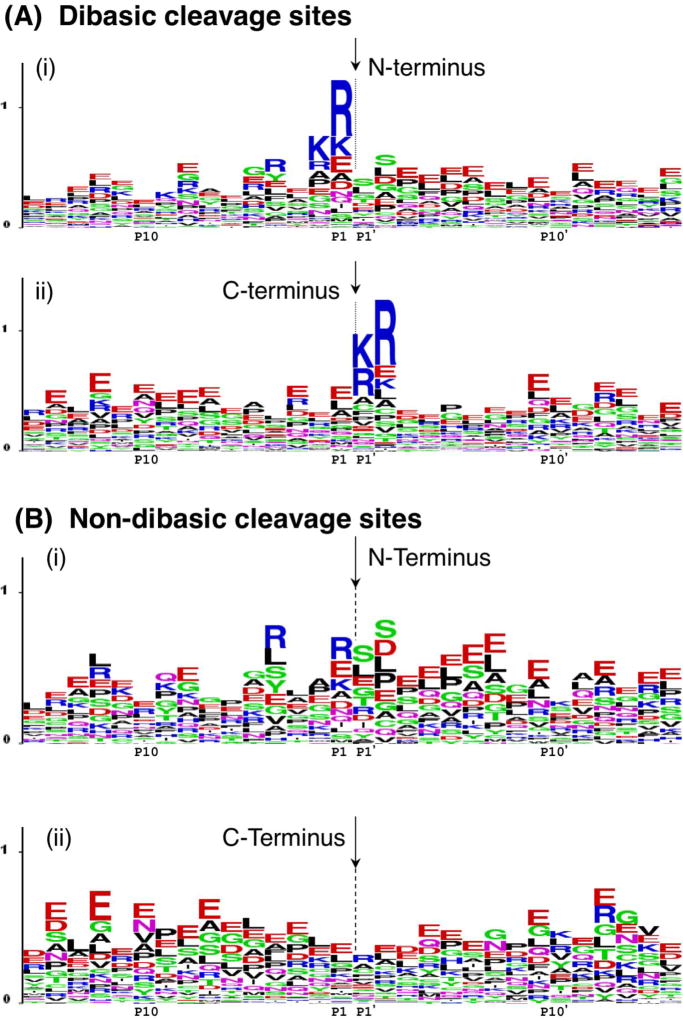
Figure 3. Pro-neuropeptide processing at dibasic and non-dibasic cleavage sites revealed by neuropeptidomics.
A. Dibasic residue processing sites. Dibasic residue sites are typical proteolytic cleavage sites of pro-neuropeptides in secretory vesicles. Sequence LOGO maps of the N- and C-termini of neuropeptides in secretory vesicles were generated by evaluation of adjacent residues to the identified peptides with their pro-neuropeptide precursors [7].
(i) N-terminal cleavage sites of identified neuropeptides. The N-termini of peptides are indicated by the arrow, with illustration of the peptide flanking amino acid sequences present within the pro-neuropeptides. The x-axis shows residues at the P1–P15 positions relative to the cleavage sites at P1-↓P1’ residues. The relative frequency of amino acids at each position is illustrated by the sequence LOGO maps.
(ii) C-terminal cleavage sites of identified neuropeptides. The C-termini of peptides are indicated by the arrow, with residues of respective pro-neuropeptides flanking the C-termini at P1’–P15’ positions.
B. Novel non-dibasic residue processing sites. Non-dibasic residue cleavage sites of pro-neuropeptides are illustrated after removal of the dibasic sites observed in the data of figure 5a.
(i) N-terminal cleavage sites. P1–P15 residues flanking the N-termini of identified peptides within their pro-neuropeptides are illustrated.
(ii) C-terminal cleavage sites. P1’–P15’ residues flanking the C-termini of identified peptides within their pro-neuropeptide precursors are illustrated.
Data for this figure was acquired as reported by Gupta et al., 2010 [7]. Briefly, dense core secretory vesicles from bovine adrenal medulla were isolated in the presence of protease inhibitors, and a low molecular weight peptide pool was prepared by millipore filtration. The sample was subjected to nano-LC-MS/MS peptidomics analyses. The identified peptides were mapped to respective pro-neuropeptide precursors to evaluate proteolytic cleavage sites. These cleavage sites were mapped by LOGO maps.