Fig. 5.
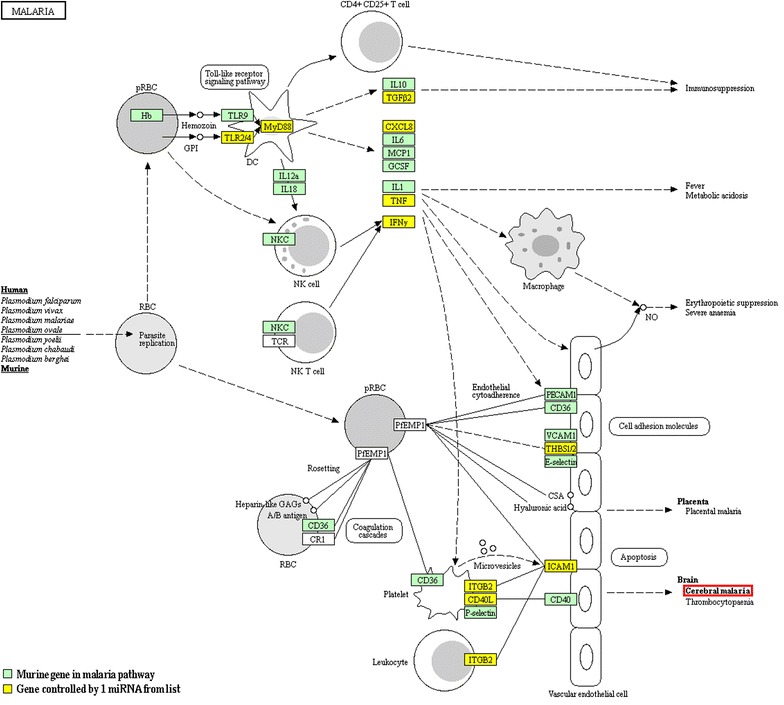
Malaria KEGG pathway. DIANA mirPath v.3 and Tarbase v.7 were used to compile lists of genes controlled by significantly differentially expressed miRNA tested using OpenArray analysis and verified by RT-qPCR analysis. Following this, the KEGG pathway for genes known to be involved in malaria infection was modified to show the role played by these miRNA. Permission has been granted for reproduction of this image. NB: some genes, including LFA1, IL8, TGFβ, and TSP are listed in the original KEGG pathway, but have been changed here to alternative but equivalent names—ITGB2, CXCL8, TGFβ2, and THBS respectively, as these gene names reflect those in the lists for the specific miRNA analyzed
