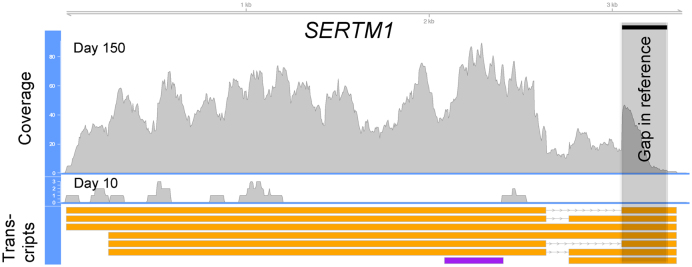
Figure 2:
Read coverage aggregated over replicate samples for the Necklace assembled superTranscript of SERTM1. This gene is found to be significantly differentially expressed using the fragments per kilobase of transcript per million mapped reads Necklace generated reference but is missed when the reference genome and annotation are used in isolation due to low read counts. The reference annotation consists of a single transcript of 321 bp (shown in purple), whereas the de novo assembled gene consists of seven transcripts up to 3,333 bp long (shown in orange) and includes approximately 250 bp that are absent from the reference genome, in a location consistent with a genome assembly gap. The genome-guided transcripts that were assembled for this gene were filtered out with StringTie's merge function due to an average fragments per kilobase of transcript per million mapped reads <1.