Figure 1.
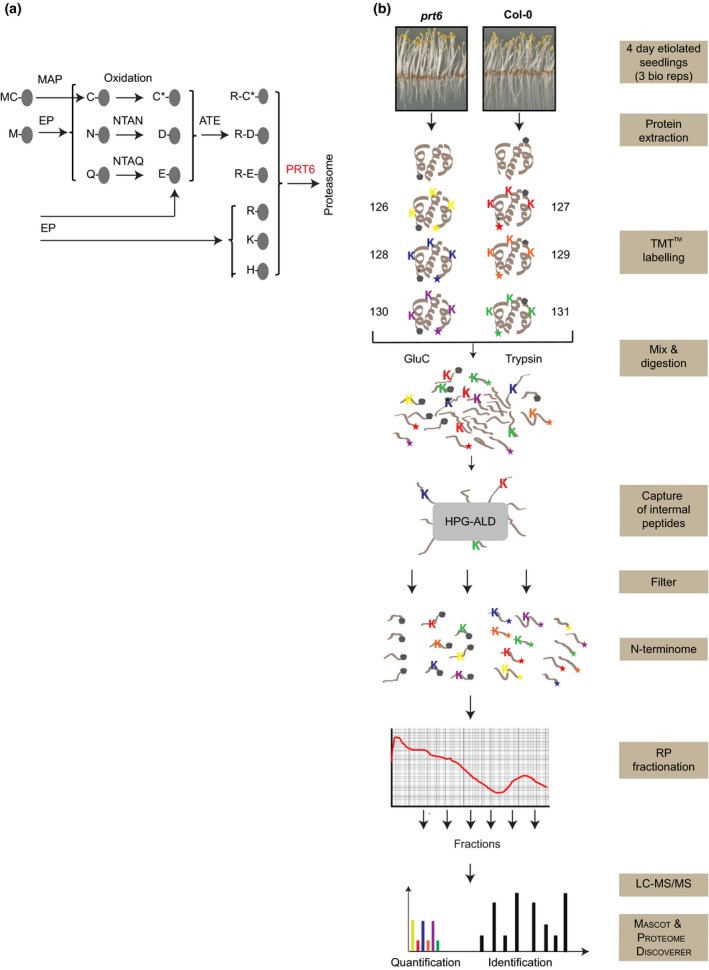
Identification and quantitation of N‐terminal peptides with TMT™‐TAILS. (a) The PRT6 branch of the Arg/N‐end rule. Substrates are generated by the action of endopeptidases (EP) or by methionine aminopeptidase (MAP)‐dependent excision of Met1 from proteins initiating Met‐Cys. PRT6, PROTEOLYSIS6 E3 ligase; ATE, arginyl tRNA transferase; NTAN1, asparagine‐specific N‐terminal amidase; NTAQ1, glutamine‐specific N‐terminal amidase. Amino acids are indicated with single letter codes; C*, oxidised cysteine. (b) Schematic representation of the TAILS workflow. Primary amines of proteins with free N‐termini (star) and lysine (K) side‐chain amines of proteins were labelled with 6‐plex TMT reagents (three biological replicates per genotype). After combining labelled samples from WT and prt6‐5 plants, the sample was divided into two, proteins were digested with either GluC or trypsin, and internal peptides were removed via hyperbranched polyglycerol aldehyde (HPG‐ALD) polymer binding of the free N‐terminal amine group. The unbound peptides (highly enriched for N‐terminal peptides) were fractionated by reversed‐phase (RP) chromatography, then analysed by high‐accuracy LC‐MS/MS. Mascot and ProteomeDiscoverer™ were used for protein identification and quantification. Grey pentagons represent naturally blocked (acetylated) N‐termini.
