Figure 4.
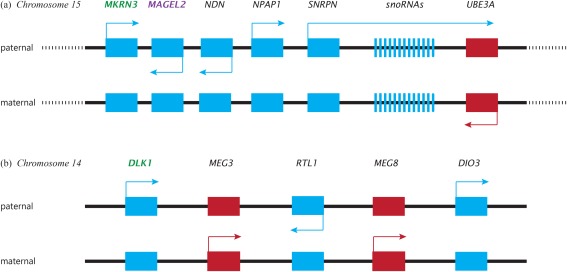
A simplified map of the cluster of imprinted genes on human chromosome 15 and 14. Red alleles are MEGs, blue alleles are PEGs. Green labels indicate known puberty promoters Purple label denotes known pubertal inhibitor. (A) Chromosome 15: The gene cluster is flanked by repeated sequences (represented by dotted lines). Recombination between the repeats in paternal meiosis results in deletion of the paternal copy of the cluster and a diagnosis of Prader–Willi syndrome. Recombination in maternal meiosis results in deletion of the maternal copy of the cluster and a diagnosis of Angelman syndrome. Mutations of the paternal copy of MKRN3 result in precocious puberty, whereas mutations of the paternal copy of MAGEL2 or deletions of the paternal cluster of small nucleolar RNAs (snoRNAs) results in hypogonadism and incomplete puberty. Mutations of the maternal copy of UBE3A are a cause of familial Angelman syndrome. (B) Chromosome 14: Temple syndrome (TS14) is caused by uniparental maternal expression of genes in this region. DLK1 mutations are associated with precocious puberty. Genetic variants in DLK1 are associated with menarche timing in girls and age at voice‐breaking in boys. Other alleles in this region are responsible for characteristics traditionally associated with kinship conflict in genomic imprinting, including maternal behaviors (MEG3/GTL2) and nonshivering thermogenesis in newborn mice (DIO3) [Color figure can be viewed at http://wileyonlinelibrary.com]
