Abstract
In most eukaryotes, including Saccharomyces cerevisiae, glycerophospholipids are the main membrane lipid constituents. Besides serving as general membrane ‘building blocks’, glycerophospholipids play an important role in determining the physical properties of the membrane, which are crucial for proper membrane function. To ensure optimal physical properties, membrane glycerophospholipid composition and synthesis are tightly regulated. This review will summarize our current knowledge of factors and processes determining the membrane glycerophospholipid composition of the reference eukaryote S. cerevisiae at the level of molecular species. Extrapolating from relevant model membrane data, we also discuss how modulation of the molecular species composition can regulate membrane physical properties.
Keywords: membrane fluidity, phospholipid molecular species, phospholipid properties
Abbreviations
DGAT, diacylglycerol acyltransferase
FAS, fatty acid synthase
GPAT, glycerol‐3‐phosphate acyltransferase
GPC, glycerophosphocholine
LPAAT, lysoPA acyltransferase
PL, phospholipid
SFA, saturated fatty acid
UFA, unsaturated fatty acid
Biological membranes are composed of a complex mixture of lipid molecules, in which proteins are embedded. Technological advances over the last decade, especially in the field of mass spectrometry, have allowed detailed analysis of the cellular lipidome, revealing thousands of different lipid molecular species 1. Furthermore, the membranes within the cell were found to differ largely in lipid composition, and the lipid composition is tightly regulated 2, 3. The latter strongly indicates that membrane lipids serve more functions than just as ‘building blocks’ of the membrane matrix. The findings that aberrant lipid composition causes endoplasmic reticulum (ER) stress and triggers stress response signaling further support the important role of lipids in membrane functioning 4. In addition, aberrant lipid metabolism has been associated with the pathology of several diseases, including the hepatic stress observed in obesity 5, fatty acid‐induced lipotoxicity 6, and the promotion of nonalcoholic fatty liver disease 7.
The proper (local) membrane lipid composition is important for organization and dynamics of membranes to allow processes such as budding, membrane trafficking, and fusion and fission events 1, the formation of domains 8, and (transient) formation of protein–protein and protein–lipid complexes. In addition, lipid composition is important for the proper embedding of membrane proteins 9, 10. The lipids surrounding a membrane protein are crucial for its structure and function, either because of direct interaction between a specific lipid and the protein 9 or because of the physical properties of the membrane matrix surrounding the protein, e.g., fluidity, thickness, intrinsic curvature, or lateral pressure profile.
The membrane lipids of the budding yeast Saccharomyces cerevisiae can be divided into three different families, based on their molecular structure: glycerophospholipids (or phospholipids in short; PL), sterols, and sphingolipids. Phospholipids make up most of the lipids of the membrane bilayer and therefore play an important role in determining the physico‐chemical properties of the membrane. This review will focus on the role of bulk phospholipids in determining membrane physico‐chemical parameters, with emphasis on the effects of acyl chain and PL class composition, and using the eukaryote baker's yeast as a reference. Research on membrane lipid housekeeping in yeast has been pivotal in the understanding of lipid homeostasis. We will first briefly introduce phospholipid biosynthesis in yeast (section Membrane phospholipid biosynthesis). Subsequently factors determining (the regulation of) membrane acyl chain composition will be discussed, including the role of substrate specificities of phospholipid biosynthetic enzymes (section Processes regulating acyl chain composition and PL molecular species profile). Next, we will review biophysical data from relevant model system studies, and discuss their implications for membrane physico‐chemical parameters (section Phospholipid molecular species profile determines physical properties of the membrane; insights from biophysical studies). Finally, we will discuss some open questions in our understanding of the regulation of membrane lipid composition in maintaining the membrane physical properties (section Conclusions and perspectives).
Membrane phospholipid biosynthesis
The phospholipid composition of yeast cells is determined by lipid metabolic processes. Lipid composition and metabolism is highly dependent on culture conditions, including temperature (section Processes regulating acyl chain composition and PL molecular species profile), growth phase 11, 12, carbon source 12, 13, the use of rich‐ or minimal media 12 and the presence of lipid precursors such as inositol 14, therefore care must be taken when comparing data from different studies.
Here, we provide a brief overview of phospholipid biosynthesis. For more general reviews regarding the biosynthesis of phospholipids in yeast and its regulation, the reader is referred to other recent reviews 15, 16, 17.
Acyl chain biosynthesis, desaturation and elongation
The bulk synthesis of acyl chains (or fatty acids) starts with acetyl‐CoA (C2) that is converted into malonyl‐CoA (C3) by the acetyl‐CoA carboxylase Acc1p, which is the rate limiting step in acyl‐CoA biosynthesis 18. Malonyl‐CoA is then used as a C2 donor by the fatty acid synthase (FAS) complex consisting of Fas1p and Fas2p, in elongating the growing acyl chain starting from acetyl‐CoA, yielding the main yeast acyl‐CoAs C16:0 and C18:0 and minor amounts of C10:0, C12:0, and C14:0 18. The acyl‐CoA elongases Elo1p, Elo2p, and Elo3p together with Ifa38p, Phs1p, and Tsc13p in the ER can elongate C14:0‐CoA and C16:0‐CoA to C18:0‐CoA and further up to C26:0 18.
C16:0‐CoA, C18:0‐CoA, and C14:0‐CoA to a minor extent can be desaturated by the Δ9‐desaturase Ole1p to C16:1Δ9‐CoA, C18:1Δ9‐CoA, and C14:1Δ9‐CoA, respectively, all of which are in cis‐conformation 19. It has been shown that C14:1Δ9‐CoA can be elongated to C16:1Δ11‐CoA 20. The subsequent Elo1p‐dependent elongation of C16:1Δ11 to C18:1Δ13 was observed when cells were grown in C14:1Δ9‐supplemented media, but even under these conditions C18:1Δ13 was only a minor species (0.5%) 20. Furthermore, C16:1Δ9‐CoA can be elongated to C18:1Δ11‐CoA, also in an Elo1p‐dependent manner 20, 21.
Triglycerides have been proposed to play a role in fatty acid detoxification by storing toxic acyl chains 22, 23. As the level of C18:1Δ11 was increased from ± 2.5% to over 10% in a mutant deficient in neutral lipid synthesis compared to the wild‐type, the elongation of C16:1Δ9‐CoA to C18:1Δ11‐CoA was suggested to be relevant for protecting against lipotoxic effects 21. This was supported by the fact that the neutral lipid deficient mutant was inviable on media containing exogenous C16:1Δ9 or C18:1Δ9, but not on media containing exogenous C18:1Δ11 21.
Glycerophospholipid biosynthesis
The precursor lipid class for all phospholipid classes in yeast is phosphatidic acid (PA), which is synthesized from the chiral water‐soluble precursor sn‐glycerol‐3‐phosphate (D‐glycerol‐1‐phosphate; G3P) in the ER (Fig. 1). The G3P acyltransferases (GPATs) Sct1p and Gpt2p acylate G3P to sn‐1‐acyl‐G3P or lysoPA using acyl‐CoA as acyl chain donor 24. Sct1p and Gpt2p can also use dihydroxyacetone phosphate (3‐hydroxy‐2‐oxopropyl phosphate; DHAP) as a substrate to produce the pro‐chiral acyl‐DHAP which is reduced to the chiral lysoPA by Ayr1p 25. Subsequently, the lysoPA acyltransferases (LPAATs) Slc1p, Ale1p, Ict1p, and Loa1p can acylate lysoPA at the sn‐2 position to form PA 26, 27, 28. As the deletion of SLC1 and ALE1 is synthetically lethal, Slc1p and Ale1p are considered the major LPAATs 26.
Figure 1.
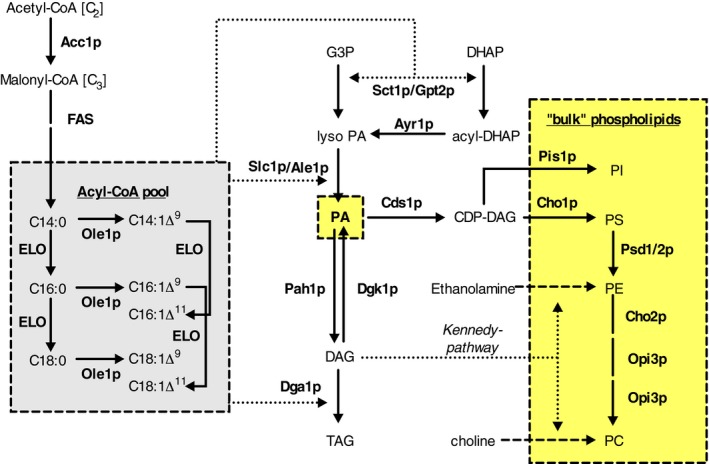
Overview of the main phospholipid biosynthetic pathways in yeast. Enzymes involved in the synthesis of bulk PLs are indicated (in bold). ELO indicates elongation processes by Elo1p and/or Elo2p. The PS‐decarboxylation step (Psd1/2p) is the only key step not localized to ER or cytosol.
PA can be either dephosphorylated to diacylglycerol (DAG) by the PA hydrolase Pah1p (in the ER) 29 or activated to CDP‐DAG by Cds1p in the ER or by Tam41p in mitochondria 30, 31. CDP‐DAG is converted into phosphatidylserine (PS) by Cho1p 32, or phosphatidylinositol (PI) by Pis1p 33, both localized in the ER. In the mitochondria, CDP‐DAG is used as precursor for the synthesis of sn‐1‐phosphatidylglycerol‐3‐phosphate by Pgs1p 34, which is dephosphorylated to phosphatidylglycerol by Gep4p 35 and subsequently combined with a second CDP‐DAG molecule by Crd1p to form the mitochondrial lipid cardiolipin (CL) 36, 37, 38. PS can be decarboxylated to phosphatidylethanolamine (PE) by the mitochondrial Psd1p or the endosomal Psd2p, both of which require transport of PS from the ER 39. The produced PE can return to the ER to be methylated by Cho2p to monomethyl‐PE, which is subsequently methylated to dimethyl‐PE and PC by Opi3p 40.
DAG is the lipid precursor for an alternative route of PE and PC synthesis via the CDP‐ethanolamine and CDP‐choline pathway, respectively, also known as the Kennedy pathway 41. In this pathway, the water‐soluble lipid head group precursors ethanolamine and choline are phosphorylated to phosphoethanolamine and phosphocholine by Eki1p and Cki1p, respectively, and subsequently activated to CDP‐ethanolamine and CDP‐choline by Ect1p and Pct1p. The CDP‐activated head group moieties can react with DAG to form PE and PC in reactions catalyzed by Ept1p and Cpt1p, respectively, which are localized in the ER and/or Golgi apparatus 42.
DAG is also the precursor for the synthesis of the storage lipid triacylglycerol (TAG) by the diacylglycerol acyltransferase (DGAT) Dga1p, which uses acyl‐CoA as acyl chain donor, or by Lro1p that uses a phospholipid as acyl chain donor 43. DAG can also be phosphorylated by the DAG kinase Dgk1p to yield PA 44.
Processes regulating acyl chain composition and PL molecular species profile
As a poikilothermic organism, yeast adapts its membrane lipid composition in response to altered growth temperatures in a process known as homeoviscous adaptation 45, 46. Upon a switch to higher growth temperature, the average acyl chain length is increased and the level of unsaturated acyl chains (UFA), mainly C16:1, decreases (Fig. 2; see also Refs 46, 47). In terms of PL molecular species, the content of monounsaturated phospholipids is gradually increased at the expense of diunsaturated phospholipids, which is accompanied by rises in the levels of the major bilayer preferring lipid classes PC and PI at the expense of PE 12. These changes are thought to be driven by the regulation of the expression of lipid biosynthetic genes and of the activity of enzymes, and the consequent alterations of fluxes through the lipid biosynthetic pathways.
Figure 2.
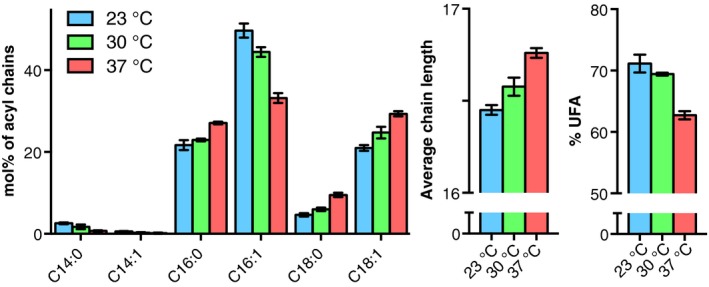
Homeoviscous adaptation in yeast: acyl chain composition at different growth temperatures. Wild‐type yeast cells (strain BY4741) were grown in synthetic defined glucose medium‐ to mid‐log phase at the indicated temperature. Total lipid extracts were prepared, transesterified to fatty acid methyl esters, and analyzed by GC‐FID (M. F. Renne and A. I. P.M. de Kroon, unpublished data). Data are presented as mean values ± SD (n = 3).
Fatty acid synthesis affects acyl chain length
The acyl chain composition of newly synthesized membrane phospholipids is mainly determined by the extent of desaturation and elongation of the acyl‐CoA pool. The length of the acyl‐CoA species in the acyl‐CoA pool may be regulated by the FAS complex, which was shown to produce different ratios of C16:0‐ to C18:0‐CoA in vitro upon temperature variation 48. In addition, the activity of the acetyl‐CoA carboxylase encoded by the ACC1 gene affects acyl chain length, as was indicated by in vitro experiments on fatty acid synthesis in which the concentration of its product malonyl‐CoA was varied 49, and further supported by a thermosensitive and a cold sensitive ACC1 mutant that shows a higher C16/C18 ratio at the nonpermissive temperature 50, 51. Recently, the regulation of the activity of Acc1p by phosphorylation has been shown to regulate the ratio of C16‐to‐C18 acyl chains in the membrane lipids 52.
Regulation of Ole1p activity
It is widely accepted that the regulation of acyl‐CoA desaturation by Ole1p plays a major role in homeoviscous adaptation in yeast. The activity of Ole1p is regulated at the level of transcription of the OLE1 gene, which is responsive to temperature, carbon source, and supplementation of exogenous fatty acids 46. The expression of OLE1 is regulated by the transcription factors Spt23p and Mga2p, which are both synthesized as 120 kDa proteins (p120) and form homodimers residing in the ER membrane 53, 54. Upon activation, Spt23p and Mga2p are ubiquitinylated and subsequently cleaved in a proteasome‐dependent manner to release 90 kDa (p90) effector proteins. The p90 proteins travel to the nucleus and facilitate OLE1 expression. The deletion of both SPT23 and MGA2 is synthetically lethal. Single spt23Δ mutants show no alterations in Ole1p levels, but mga2Δ have lower Ole1p levels and therefore lower levels of UFA chains implicating Mga2p as the major player in Ole1p regulation 55.
Recently Mga2p dimers were proposed to sense lipid packing via their transmembrane helices 56. The authors propose the aromatic residues on the transmembrane helices to face ‘outwards’ to the lipid acyl chains in a fluid environment, and rotate ‘inwards’ towards the monomer–monomer interface in a more rigid environment, due to the tighter packing of the lipid acyl chains, which disfavors the insertion of the aromatic residues into the acyl chain region. When the membrane is fluid the ubiquitination sites of the Mga2p dimer are sequestered. When the membrane lipids are tightly packed, the rotation mechanism exposes the ubiquitination site on both Mga2p monomers, enabling processing by the proteasome and release of p90 56. A similar mechanism likely applies to Spt23p, as the transmembrane helices of Spt23p and Mga2p have 86% sequence similarity 57.
Substrate specificity of acyltransferases
The substrate specificity of lipid biosynthetic enzymes determines the molecular species profile of synthesized lipids and may have a role in determining the phospholipid molecular species profile (see section Substrate specificity of PL biosynthetic pathways). Pulse labeling strategies in various yeast mutants, in vitro enzyme assays, and changes in the lipidome of deletion and overexpression mutants have been used to deduce substrate specificities at the acyl chain level.
Whereas the GPAT Gpt2p was shown to have little substrate preference in vitro (except for slightly reduced activity towards C18:0‐CoA), its paralog Sct1p was shown to exhibit a strong preference for C16:0‐CoA and C16:1‐CoA 24. In vivo, the deletion of SCT1 was shown to decrease the C16:0 levels in the four major phospholipids PC, PE, PI, and PS, with the largest effect on PC. Interestingly, deletion of GPT2 hardly affects the acyl chain composition of PC, PI, and PS, but does increase the C16:0 and C18:1 levels in PE at the expense of C16:1 24. Looking at the total cellular acyl chain composition, De Smet et al. 58 showed that deletion of SCT1 decreases the cellular C16:0 content by almost 50%, whereas overexpression of SCT1 increases C16:0 levels at the expense of C16:1 and C18:1. Based on these data, it has been hypothesized that Sct1p‐mediated incorporation of C16:0 into PL serves as a sink for C16:0. In line with this hypothesis it was shown that co‐overexpression of OLE1 rescues the growth defect conferred by SCT1 overexpression and the accumulation of C16:0, indicating competition between Sct1p and Ole1p for their shared substrate C16:0‐CoA 58. Interestingly, overexpression of OLE1 increases the level of C18:1 rather than C16:1 58, indicating a preference for C18:0‐ over C16:0‐CoA.
Loss of the LPAAT Ale1p does not significantly affect the PL molecular species profile. In contrast, loss of Slc1p causes a general increase in C32 PL at the expense of C34 PL 26. A later study showed that this is mainly due to a decrease in C18:1 acyl chains in the PL and an increase in C16:1 with only minor effects on the saturated acyl chains 59, indicating that Slc1p‐mediated incorporation of C18:1‐CoA into PLs may serve as a sink for C18:1.
In a lipidomics study using four double mutants with deletions of SCT1 or GPT2 combined with deletion of SLC1 or ALE1, it was shown that deletion of SLC1 in combination with the deletion of either SCT1 or GPT2 has a large effect on the molecular species composition, whereas deletion of ALE1 in combination with SCT1 or GPT2 shows only minor differences 60. As Ale1p, but not Slc1p, is known to have general lysoPL acyltransferase activity 61, 62, 63, 64, these findings were interpreted as indicative for de novo synthesis of PL having a more pronounced role in determining the acyl chain at the sn‐2 position 60. However, the proposed acyl‐CoA preference of Slc1p versus the absence of a clear‐cut acyl‐CoA substrate specificity for Ale1p may also contribute.
The DGAT Dga1p has been proposed to have a substrate specificity for C16:0 and C18:1, based on the observation that yeast cells lacking DGA1 show a decrease in C16:0 and C18:1 levels which is compensated by an increase in C16:1 65. Care must be taken when interpreting these data, as Dga1p was studied in a background strain deleted for the other neutral lipid synthesis genes (lro1Δare1Δare2Δ). Interestingly it was found that in the quadruple mutant dga1Δlro1Δare1Δare2Δ the total level of C18:1 is increased compared to wild‐type, which is mainly due to the presence of 7.3% C18:1Δ11 21, 65. The presence of ARE1 does not decrease the level of C18:1Δ11 and LRO1 or ARE2 only decrease it to 3.3–3.4%. However, the presence of DGA1 reduces the C18:1Δ11 level to wild‐type (0.1%), consistent with Dga1p being responsible for bulk TAG synthesis.
Besides the direct role in determining the nature of the acyl chains of the synthesized lipid, the substrate specificities of the acyltransferases described above may affect lipid biosynthesis indirectly, by their impact on the composition of the acyl‐CoA pool. By transferring the acyl chains from specific acyl‐CoA species, the acyl‐CoA substrate is no longer accessible to other enzymes catalyzing, e.g., elongation or desaturation. Variation in the activity of enzymes that use bulk amounts of specific acyl‐CoAs, such as Sct1p consuming C16:0‐CoA above, will affect sinks for acyl chains and alter the available acyl‐CoA species by affecting reactions involving acyl‐CoA's by mass action 16. In this context the acyl‐CoA synthetases Faa1‐4p and Fat1p should be mentioned that contribute to the acyl‐CoA pool by converting free fatty acids mobilized from neutral lipids or derived from lipid turnover into acyl‐CoA's 66, 67. Differences in substrate specificity both in vitro and in vivo have been reported for acyl‐CoA synthetases (reviewed in Ref. 66). The activity and specificity of the acyl‐CoA synthetases may contribute to the composition of the acyl‐CoA pool and thereby influence acyl‐CoA‐dependent metabolic steps.
Substrate specificity of PL biosynthetic pathways
The major yeast lipids PC, PE, PI, and PS have different molecular species profiles (Fig. 3, 12). For PC and PE, 32:2 and 34:2 are the most abundant together amounting to over 75%, followed by 32:1 and 34:1. Interestingly, the disaturated species 32:0 and 34:0 are not detected. The major PS species is 34:1, followed by 32:1 and 34:2. PI is enriched in the disaturated, short acyl chain species 26:0, 28:0, and 30:0 (approx. 17% of total) compared to PC, PE, and PS, in which these are absent. Furthermore, diunsaturated species are virtually absent in PI (less than 5%). The substrate use of the lipid biosynthetic pathways could contribute to the PL‐specific species profiles. The substrate use is determined by the substrate specificity of the individual enzymes and/or the molecular species of the lipid substrate available at their subcellular location. Postsynthetic acyl chain remodeling may further contribute (section Acyl chain remodeling).
Figure 3.
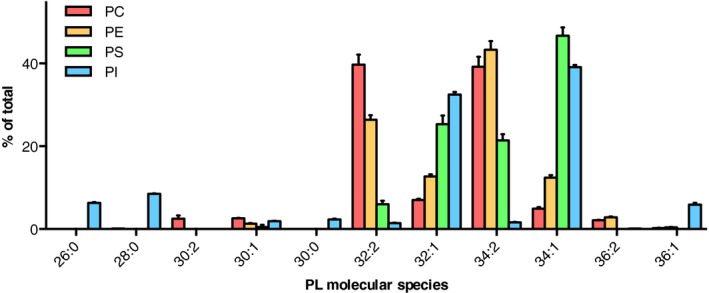
Molecular species profiles of the major membrane phospholipids in yeast. The molecular species of the main glycerophospholipids PC, PE, PS, and PI are shown of wild‐type strain BY4741, cultured to early log phase in synthetic complete glucose medium at 30 °C. Data (mean values with standard deviation; n = 3) were taken from the supplementary data of Klose et al. 12.
Substrate specificity at the level of molecular species of the PA consuming enzymes Cds1p and Pah1p, which produce CDP‐DAG and DAG, respectively, has not (yet) been reported. This also applies to the PI and PS biosynthetic enzymes Pis1p and Cho1p. However, the routes producing the bulk membrane phospholipids PE and PC were reported to show selectivity at the level of molecular species. In the absence of ethanolamine and choline, yeast is mainly dependent on PS decarboxylation for PE synthesis and subsequent methylation to PC. The mitochondrial enzyme Psd1p is the main PS decarboxylase and produces 70–90% of cellular PE 68. Compared to PS, PE and PC are enriched in diunsaturated species (Fig. 3, 12, 69, 70) which could be due to molecular species selectivity of the PS decarboxylase. Alternatively, the species selective conversion of PS to PE could be due to preferential transport of diunsaturated PS species from the ER to mitochondria, as was reported in mammalian cells 71.
The molecular species profiles of PC synthesized via either the methylation of PE by Cho2p and Opi3p or by the Kennedy pathway were shown to differ in a stable isotope pulse labeling study using D13‐choline and D3‐methionine 72. The PE methylation pathway produces mainly 34:2 and 32:2 PC species (approx. 55% and 35% of total, respectively), whereas the CDP‐choline pathway produces a more even distribution of 32:2, 32:1, 34:2, and 34:1 PC species (approx. 40%, 20%, 20%, and 10% of total, respectively). When the Kennedy pathway is inactivated, e.g., by deletion of PCT1, the wild‐type PC molecular species profile cannot be produced by biosynthesis alone, and requires acyl chain remodeling of the PE methylation‐derived PC (see section below).
Acyl chain remodeling
Recently, we reviewed the current knowledge of lipid acyl chain remodeling in yeast 73. Here, we will briefly discuss how the remodeling of PC ensures the proper molecular species profile of this bulk lipid.
PC can be de‐acylated by a phospholipase A to lysoPC, which can subsequently be de‐acylated to glycerophosphocholine (GPC) by a phospholipase B. PC can also be directly de‐acylated to GPC by a phospholipase B. GPC can either be catabolized to choline and glycerol‐3‐phosphate 74, 75, or re‐acylated to yield 1‐acyl‐glycerophosphocholine (lysoPC) 76, e.g., by the recently identified Gpc1p 77. LysoPC can be acylated to PC by a second acyltransferase such as Ale1p 62. The phospholipase D Spo14 can release the head group of PC, yielding PA and choline, which can subsequently be recycled into PC via the Kennedy pathway 78.
The de‐acylation of PC and subsequent re‐acylation of lysoPC or GPC was first demonstrated in vivo by Wagner & Paltauf using the incorporation of exogenous radiolabeled fatty acids 79. The incorporation of acyl chains was shown to be a very rapid process, observed already after 2 min, however, the incorporation of the label in the lipid pools was not quantified. Using a strain deficient in PE methylation (cho2∆opi3∆) with a switchable Kennedy pathway (P GAL1 ‐PCT1) it was shown that cells can survive on exogenous diC8‐PC in the absence of active PC synthesis. Moreover, diC8‐PC was shown to be remodeled to the regular PC species such as 32:2, 32:1, 34:2, and 34:1. However, cell growth is delayed and the content of PC is diminished to less than 10% of total compared to over 40% in the wild‐type, indicating that the uptake of diC8‐PC followed by PC remodeling is not sufficiently efficient to sustain proper PC levels and optimal cell growth 80.
As mentioned above, in the pct1Δ background newly synthesized PC is mainly diunsaturated as in wild‐type and contains virtually no monounsaturated species. However, the steady‐state PC profile does contain monounsaturated species 72 indicating a role for remodeling of PC. This was further supported by the finding that the sn‐1 position of newly synthesized PC contains almost exclusively C16:1, whereas steady‐state PC contains C16:0, C16:1, C18:0, or C18:1 at the sn‐1 position 72. A follow‐up pulse‐chase study in the pct1∆ background using the overexpression of SCT1 to increase the level of saturated acyl chains, revealed that the monounsaturated PC species are increased to nearly steady‐state levels at the expense of diunsaturated PC species in approximately 1 doubling time, involving approximately 40% of the newly synthesized PC molecular species 81. Deletion of the phospholipase B encoding gene PLB1 was shown to slow down this process, indicating a remodeling mechanism via the turnover of PC to GPC. Subsequent acylation of GPC to lysoPC by Gpc1p 77, followed by acylation to PC by Ale1p 76, 82 are likely major contributors to this process.
Phospholipid molecular species profile determines physical properties of the membrane; insights from biophysical studies
Understanding how physico‐chemical properties of membrane lipids affect membrane properties is key to understanding the importance of the phospholipid molecular species profile for maintaining membrane function in vivo. Biophysical studies on model membranes consisting of (mixtures of) well‐defined synthetic lipids or purified lipid extracts have provided much information on how lipid physico‐chemical properties determine membrane physical parameters such as fluidity, bilayer thickness, intrinsic curvature, and lateral pressure profile. It is important to note that most of these parameters are interrelated and therefore a single parameter cannot be modified without affecting the other parameters to some extent. Nevertheless, these studies have enormously contributed to our understanding of the role of lipids in the complex environment of a biological membrane. In the following, we will focus on the various ways in which the phospholipid molecular species profile can affect membrane physical properties and we will discuss possible implications with respect to regulation of membrane lipid composition in vivo.
Membrane fluidity
Membrane fluidity is probably the most extensively studied physical property of membranes, both in vitro and in vivo. Depending on the temperature, a lipid bilayer can exist in different phases. The two major, biologically relevant phases are the liquid crystalline phase (L α), a fluid and disordered phase, and the gel phase (L β), which forms at lower temperature and is a more ordered and rigid phase. The phase transition temperature (T m) from the gel to the liquid crystalline phase has been determined for many different lipids, using model membranes composed of pure lipid molecular species.
Role of acyl chain composition
The fluidity of a membrane at a given temperature is mainly determined by the acyl chain composition 83. The T m increases with increasing lipid acyl chain length 84 and decreases with the introduction of cis‐double bonds in the acyl chains 85 (Fig. 4). The decrease in T m is larger going from a completely saturated lipid species to a monounsaturated species than from a mono‐ to diunsaturated species (Fig. 4, compare grey arrows. For unsaturated PL species, the location of the cis‐double bond also affects T m (Fig. 5). Yeast acyl chains comprise mainly Δ9 UFAs with only minor amounts of Δ11 UFAs (see section Acyl chain biosynthesis, desaturation and elongation; 21, 65). As the difference in T m between Δ9 and Δ11 PL species is very small, this most likely will play but a minor role – if any.
Figure 4.
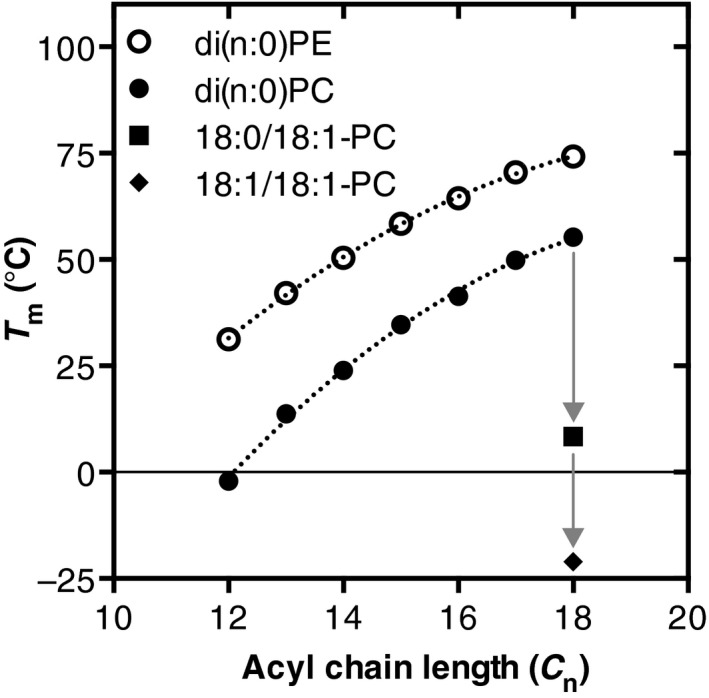
Effect of acyl chain length and unsaturation on the gel‐to‐liquid crystalline phase transition temperature (T m) of PC and PE. T m of completely saturated di(n:0), monounsaturated (n:0)(n:1) and diunsaturated di(n:1) PE and PC species with acyl chains containing the same number of carbons, n. Grey arrows highlight the differences in T m between saturated, monounsaturated, and diunsaturated PC species. Data were taken from 84 for di(n:0)PC, 112 for (n:0)(n:1)PC, 113 for di(n:1)PC, and 114 for di(n:0)PE. All data were obtained by differential scanning calorimetry of lipid dispersions prepared in water. Curves were added to guide the eye.
Figure 5.
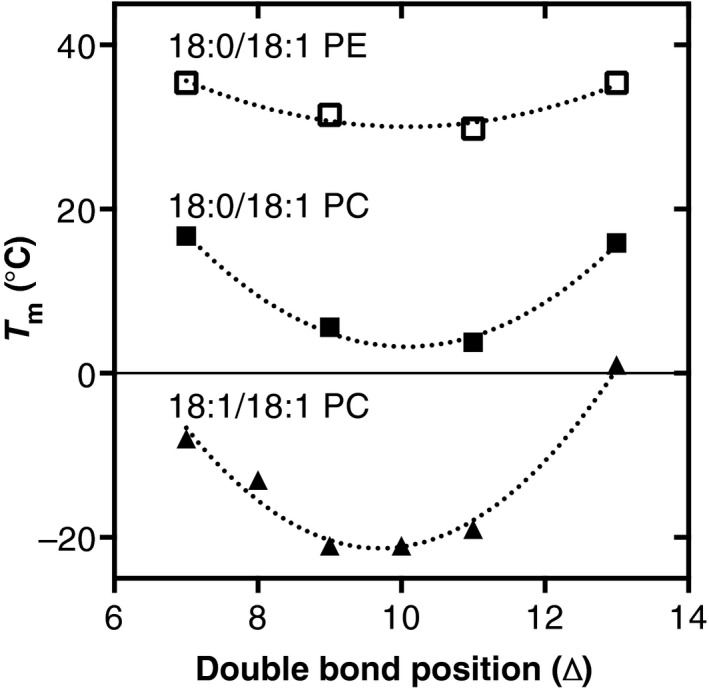
The effect of the position of the cis‐double bond (Δ‐location) on the gel‐to‐liquid crystalline phase transition (T m) of PE and PC species. All data were obtained by differential scanning calorimetry. Representative data were taken from 113 for 18:1/18:1 PC, from 115 for 18:0/18:1 PC, and 116 for 18:0/18:1 PE. Curves were fitted to guide the eye.
The fluid phase is thought to represent the bulk of the membrane lipids. Maintaining the fluidity of the membrane at altered temperatures is accomplished by adaptation of the acyl chain composition in both degree of unsaturation and acyl chain length (Fig. 2, see also section Processes regulating acyl chain composition and PL molecular species profile). The decreased acyl chain length and increased unsaturation observed at lower growth temperatures are in line with the lower T m of shorter and more unsaturated PL species as determined in model membranes (Fig. 4).
Role of lipid class composition
Membrane fluidity is also dependent on lipid class composition. PE has a smaller head group than PC and can form hydrogen bonds between the positively charged amino head group and the negatively charged phosphate residue of a neighboring lipid 86. Both these properties cause tighter packing of PE and its acyl chains, which leads to a higher T m of PE molecular species compared to the corresponding PC species (Fig. 4). Recently, it was shown that the PE‐to‐PC ratio can be adapted to maintain membrane fluidity in insect cells as well as mammalian cells upon varying the cholesterol content 87. Interestingly, yeast cells grown at higher temperature show a decrease in PE content 12, which is in contrast to the higher T m of PE compared to PC (Fig. 4). Therefore, this decrease in PE content is likely to balance membrane intrinsic curvature and lateral pressure profile (see below).
Difference in effects between sn‐regioisomers of phospholipid species
Lipid species are denoted in various ways, depending on the level of molecular species structural information 88. For example, lipids denoted as 32:1 PC species (defined as a PC lipid with two acyl chains of total 32 C atoms and one double bond) comprise many possible molecular species depending on the acyl chain composition and the sn‐regioisomers, for example: 14:0/18:1PC versus 18:1/14:0PC, where the first fatty acid is esterfied to the sn‐1 position and the second one to the sn‐2 position on the glycerol backbone of PC 88. The effect of lipid sn‐regioisomers on the total molecular species composition in yeast has not received much attention so far. However, biophysical studies using model membranes have shown striking differences between sn‐regioisomers, e.g., in their melting temperature (Table 1). Differences in the insertion depth of the sn‐1 and sn‐2 acyl chains have been shown, where the sn‐2 inserts less deeply into the bilayer, leading to different effects of the sn‐regioisomers on lipid packing 89.
Table 1.
Effect of sn‐regioisomers on the gel‐to‐liquid crystalline phase transition temperature (T m). Data were taken from The Handbook of Lipid Bilayers (second edition, 2013) and references therein 120. Tm of lipids with the same acyl chain at the sn‐1 and sn‐2 positions are depicted in bold
| sn‐2 | |||||
|---|---|---|---|---|---|
| 12:0 | 14:0 | 16:0 | 18:0 | 18:1 | |
| sn‐1 | |||||
| 12:0 | −2.1 | 21.7 | 23.3 | ||
| 14:0 | 23.9 | 34.9 | 39.2 | −19.1 | |
| 16:0 | 11.3 | 27.5 | 41.4 | 48.8 | −4 |
| 18:0 | 17.5 | 31 | 44.5 | 55.3 | 5.6 |
| 18:1 | −9 | 10 | −17.3 | ||
For monounsaturated lipids, in general, the regioisomer with the saturated or longer acyl chain at the sn‐1 position has a lower melting temperature compared to the regioisomer with the saturated or longer acyl chain at the sn‐2 position. This might explain the strong enrichment of saturated acyl chains at the sn‐1 position in yeast PL 79 that may serve as a mechanism to mitigate the effect of these acyl chains on membrane rigidity. To our knowledge, no studies have investigated the regulation of sn‐regioisomers in relationship with the regulation of membrane fluidity in yeast.
Influence of sterols
The addition of sterols to a phospholipid bilayer diminishes the differences between the fluid and gel phases. By ordering the acyl chains in a liquid crystalline phase, sterols make membranes more rigid. Conversely, in the gel phase, sterols cause disorder of the saturated acyl chains, thus increasing membrane fluidity. Because of decreased differences between the gel phase and liquid crystalline phase at high sterol content, the phase transition is no longer observed 90. In yeast, ergosterol content was shown to differ only slightly in yeast cultures grown at different temperatures 12.
Membrane thickness
Membrane thickness is determined by the effective length of the lipid acyl chains, which is mainly determined by the number of carbon atoms in the acyl chain but can be modulated by several factors. First, the bilayer thickness of a lipid in a gel phase will be more ordered than in a fluid phase, and hence the bilayer will be thinner in a fluid phase membrane. Similarly, bilayer thickness of a membrane composed of unsaturated lipids will be slightly less than that of a bilayer consisting of the corresponding saturated lipid. Furthermore, for lipids in the fluid phase, bilayer thickness will decrease with increasing temperature due to increased acyl chain disorder. Consistently, sterols generally increase membrane thickness because of their stretching/ordering effects on especially saturated acyl chains in the fluid phase.
In yeast cells the plasma membrane is the thickest membrane and accordingly it is enriched in sterols and saturated lipid species 2, 70. Consequently, plasma membrane‐localized proteins have significantly longer transmembrane segments than Golgi or ER resident membrane proteins 91. Matching of transmembrane helix length to the bilayer thickness (hydrophobic matching) has been proposed as a mechanism for sorting and retention of proteins 92.
The close relationship between bilayer thickness and membrane fluidity is reflected in the mechanism proposed for the regulation of membrane fluidity by the bacterial thermosensor DesK 93, 94. When bacteria are exposed to lower temperatures, DesK is activated and downstream signaling leads to expression of a desaturase which increases membrane lipid unsaturation 94. DesK has been proposed to sense the temperature‐induced decrease in membrane fluidity as an increase in membrane thickness, based on studies using a “minimal sensor” mutant of DesK 93. Recently, the yeast ER stress sensor Ire1p was proposed to sense the physical state of the membrane by sensing membrane thickness 95. The authors present evidence that Ire1p locally compresses the membrane bilayer. The higher energetic cost of bilayer compression resulting from increased bilayer thickness induced by aberrant lipid composition is proposed to drive Ire1p dimerization 95.
Lipid polymorphism and membrane intrinsic curvature
Not all phospholipid species, when hydrated, assemble in bilayers. Indeed, there are many examples of lipids that have a preference for assembling as nonbilayer phases, e.g., an inverted hexagonal phase or micelle. The phase preference of a lipid can be rationalized by the simple, but effective ‘shape structure’ concept 96, in which the ratio of the cross‐sectional area of the lipid head group over the cross‐sectional area of the acyl chain region determines which phase the lipids adopt. When the cross‐sectional area of the lipid head group is close to that of the acyl chain region, the molecule is cylindrical and prefers to form bilayers. In type I lipids, such as various lysoPLs, the head group cross‐sectional area is large compared to that of the acyl chain, giving them the tendency to form positively curved structures. Therefore, type I lipids assemble as micelles in aqueous environment. Conversely, type II lipids, e.g., most diunsaturated PE and PA molecular species, preferably form inverted hexagonal phases upon hydration due to their small head group area compared to that of the acyl chains 97. It should be realized that the nature of the acyl chains determines the extent of the nonbilayer preference or intrinsic curvature by influencing the shape of the molecule 98. This is reflected in the bilayer‐to‐hexagonal (H II) phase transition temperature (T H; Fig. 6).
Figure 6.
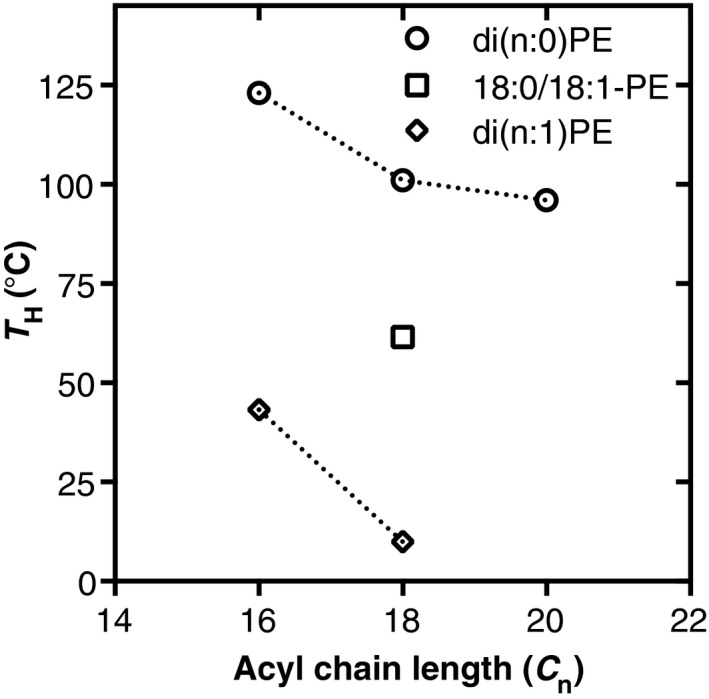
Effect of acyl chain composition on the bilayer‐to‐hexagonal phase transition temperature (TH) of PE. All data were obtained by differential scanning calorimetry. Representative data were taken from 117 for di(n:0)PE, 118 for 18:0/18:1‐PE, and 119 for di(n:1)PE.
A lipid monolayer of type II nonbilayer preferring lipids will have the tendency to form a negatively curved surface (i.e., intrinsic curvature). When mixed with bilayer lipids, type II lipids can be forced to assemble in a bilayer allowing, e.g., mitochondrial membranes to form bilayers despite their very high content of the nonbilayer lipids PE and CL. However, due to the intrinsic curvature of type II lipids, this will induce curvature stress 97. The presence of nonbilayer lipids and possibly the transient formation of nonbilayer structures have been proposed to be important for fusion and fission processes 99.
Yeast cells were proposed to regulate the intrinsic curvature, by modulating the PE molecular species profile 100. Upon depletion of PC in a yeast cho2opi3 mutant, PE levels increase and PE takes over as the major membrane PL. In parallel, the proportion of monounsaturated PE species increases at the expense of diunsaturated species, and a shortening of the average acyl chain length is observed. As diunsaturated PE species and PE species with longer acyl chains have stronger nonbilayer propensity, as shown by a lower T H (Fig. 6), the decrease in these species was interpreted as a mechanism to regulate membrane intrinsic curvature in response to PC depletion. Indeed, PE extracted from yeast cells under PC depleting conditions was found to have a higher T H, indicative for PE species that are more bilayer compatible. The accompanying shortening of the average chain length of the PE molecular species also decreases the T m of the PE, consistent with the adaptation of the PE molecular species also compensating for the decreased membrane fluidity that would result from the increased PE level and decrease in unsaturated PE species (Fig. 4).
Lateral pressure profile
The intrinsic curvature of a membrane is closely related to the lateral pressure profile of a membrane, arising from the same structural properties of the membrane lipids 99. The lateral pressure profile reflects the depth‐dependent distribution of lateral stress in a membrane 101. The nature of this phenomenon and its relevance for membrane structure and function can be qualitatively understood as follows. The hydrophobic effect causes lipids to come together and form bilayers, giving rise to an attractive force, or negative lateral pressure, between the lipids at the head group–acyl chain interface. At the same time, interactions between neighboring acyl chains in the hydrophobic core and between hydrophilic head groups with bound water molecules will cause steric repulsion, giving rise to a positive lateral pressure in these regions. Together this leads to an inhomogeneous pressure profile along the bilayer normal (i.e., the lateral pressure profile). Importantly, the lateral pressure is net zero in self‐assembled bilayer structures 102.
Upon increasing the ratio of nonbilayer‐to‐bilayer preferring lipids, the (positive) lateral pressure between the head groups is reduced because of the smaller effective head group volume of the nonbilayer lipids. As a consequence, the lipids will pack more tightly, leading to an increase in the lateral pressure between the acyl chains.
The lateral pressure profile of a membrane affects membrane protein structure and stability 103, 104. As the membrane intrinsic curvature and the lateral pressure profile are related properties, the lateral pressure profile is likely to be regulated in a similar manner as the intrinsic curvature and this may affect protein conformation and/or function 105. For example, a protein in a membrane with a high PE content will have more freedom of movement in the head group area due to the locally lower lateral pressure, but will experience higher lateral pressure in the hydrophobic core 99. In addition, the presence of PE with its relatively small head group may promote insertion of hydrophobic molecules into the lipid/water interface 106. Vice versa, insertion of hydrophobic molecules into a bilayer may modulate the lateral pressure profile. For example, the insertion of alcohols in the head group region of a membrane has been suggested to destabilize membrane proteins by increasing the lateral pressure in the head group region and decreasing the lateral pressure in the hydrophobic core 106.
Conclusions and perspectives
Thorough genetic studies in combination with lipid analysis and lipidomics studies have provided insight into phospholipid biosynthesis and its regulation. However, mechanisms such as the regulation of acyl chain remodeling, the sensing of membrane physico‐chemical properties, and its downstream signaling to the lipid biosynthetic machinery largely remain to be elucidated. Recently, the molecular mechanisms of membrane sensing by the OLE1 transcriptional regulator Mga2p and the ER stress sensor Ire1p have been elucidated 56, 95, as described above. Whereas Mga2p responds to differences in lateral pressure, Ire1p is proposed to sense bilayer thickness by a mechanism based on membrane compression that is dependent on the amphipathic helix neighboring the transmembrane helix. Membrane binding amphipathic helices occur in many proteins and have been well studied in terms of the membrane physical parameters they recognize and how membrane binding influences their function 107, 108. Furthermore, membrane binding by amphipathic helices can be modulated, e.g., the amphipathic helix of Pah1p is not recruited to PA‐containing membranes upon phosphorylation 109.
Although transcriptional regulation via the Henry regulatory circuit controlled by Opi1p sensing the PA level is well characterized for a set of lipid biosynthetic genes containing UASINO in their promoter 15, regulation at the (post‐)translational level of most lipid biosynthetic enzymes is not understood, with the notable exception of Pah1p 29. Many lipid biosynthetic enzymes are phosphorylated 110. However, the function of phosphorylation in the regulation of, e.g., activity, subcellular localization or degradation is often not known. Furthermore, the kinases and phosphatases involved in the phosphorylation remain to be identified for most enzymes.
Recent advances in lipidome analysis have provided opportunities to identify lipid characteristics that were previously difficult to distinguish, such as double bond localization and sn‐regioisomers 111. Integration of these state‐of‐the‐art lipid analysis techniques with stable isotope labeling will enable dynamic lipidomics studies that will help reveal mechanisms in the regulation of PL molecular species profiles. By combining observations from such experiments with current and new data from biophysical studies on synthetic lipids and biological lipid extracts, this approach will provide novel insights into the regulation of key physico‐chemical properties of the membrane such as membrane fluidity.
Acknowledgements
We gratefully acknowledge Prof. Dr. Antoinette Killian for critical reading of the manuscript and expert input. Furthermore, we thank the current and former members of the yeast lipid group of the Membrane Biochemistry & Biophysics section for fruitful discussions. Our work is supported by the Division of Chemical Sciences in the Netherlands, with financial aid from The Netherlands Organization for Scientific Research (NWO).
Edited by Wilhelm Just
References
- 1. van Meer G, Voelker DR and Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9, 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Meer G and de Kroon AIPM (2011) Lipid map of the mammalian cell. J Cell Sci 124, 5–8. [DOI] [PubMed] [Google Scholar]
- 3. Barelli H and Antonny B (2016) Lipid unsaturation and organelle dynamics. Curr Opin Cell Biol 41, 25–32. [DOI] [PubMed] [Google Scholar]
- 4. Volmer R and Ron D (2015) Lipid‐dependent regulation of the unfolded protein response. Curr Opin Cell Biol 33, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fu S, Yang L, Li P, Hofmann O, Dicker L, Hide W, Lin X, Watkins SM, Ivanov AR and Hotamisligil GS (2011) Aberrant lipid metabolism disrupts calcium homeostasis causing liver endoplasmic reticulum stress in obesity. Nature 473, 528–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kohlwein SD (2010) Obese and anorexic yeasts: experimental models to understand the metabolic syndrome and lipotoxicity. Biochim Biophys Acta 1801, 222–229. [DOI] [PubMed] [Google Scholar]
- 7. Leamy AK, Egnatchik RA and Young JD (2013) Molecular mechanisms and the role of saturated fatty acids in the progression of non‐alcoholic fatty liver disease. Prog Lipid Res 52, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Simons K and Toomre D (2000) Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 9. Dowhan W, Bogdanov M and Mileykovskaya E (2016) Functional roles of lipids in membranes In Biochemistry of Lipids, Lipoproteins and Membranes (Ridgway N. and McLeod R, eds), pp. 1–40. Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 10. Nilsson I, Ohvo‐Rekilä H, Slotte JP, Johnson AE and von Heijne G (2001) Inhibition of protein translocation across the endoplasmic reticulum membrane by sterols. J Biol Chem 276, 41748–41754. [DOI] [PubMed] [Google Scholar]
- 11. Janssen MJFW, Koorengevel MC, De Kruijff B and De Kroon AIPM (2000) The phosphatidylcholine to phosphatidylethanolamine ratio of Saccharomyces cerevisiae varies with the growth phase. Yeast 16, 641–650. [DOI] [PubMed] [Google Scholar]
- 12. Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A and Simons K (2012) Flexibility of a eukaryotic lipidome – insights from yeast lipidomics. PLoS One 7, e35063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tuller G, Nemec T, Hrastnik C and Daum G (1999) Lipid composition of subcellular membranes of an FY1679‐derived haploid yeast wild‐type strain grown on different carbon sources. Yeast 15, 1555–1564. [DOI] [PubMed] [Google Scholar]
- 14. Gaspar ML, Aregullin MA, Jesch SA and Henry SA (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae . J Biol Chem 281, 22773–22785. [DOI] [PubMed] [Google Scholar]
- 15. Henry SA, Kohlwein SD and Carman GM (2012) Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae . Genetics 190, 317–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. De Kroon AIPM, Rijken PJ and De Smet CH (2013) Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog Lipid Res 52, 374–394. [DOI] [PubMed] [Google Scholar]
- 17. Carman GM and Han G‐SS (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae . Annu Rev Biochem 80, 859–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tehlivets O, Scheuringer K and Kohlwein SD (2007) Fatty acid synthesis and elongation in yeast. Biochim Biophys Acta 1771, 255–270. [DOI] [PubMed] [Google Scholar]
- 19. Stukey JE, McDonough VM and Martin CE (1989) Isolation and characterization of OLE1, a gene affecting fatty‐acid desaturation from Saccharomyces cerevisiae . J Biol Chem 264, 16537–16544. [PubMed] [Google Scholar]
- 20. Schneiter R, Tatzer V, Gogg G, Leitner E and Kohlwein SD (2000) Elo1p‐dependent carboxy‐terminal elongation of C14:1Delta(9) to C16:1Delta(11) fatty acids in Saccharomyces cerevisiae . J Bacteriol 182, 3655–3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sec P, Garaiova M, Gajdos P, Certik M, Griac P, Hapala I and Holic R (2015) Baker's yeast deficient in storage lipid synthesis uses cis‐vaccenic acid to reduce unsaturated fatty acid toxicity. Lipids 50, 621–630. [DOI] [PubMed] [Google Scholar]
- 22. Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D'Ambrosio D, Ruggles KV, Ramsey N, Jabado O, Turkish A and Sturley SL (2009) Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid‐mediated cell death. J Biol Chem 284, 30994–31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petschnigg J, Wolinski H, Kolb D, Zelling G, Kurat CF, Natter K and Kohlwein SD (2009) Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem 284, 30981–30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zheng Z and Zou J (2001) The initial step of the glycerolipid pathway: identification of glycerol 3‐phosphate/dihydroxyacetone phosphate dual substrate acyltransferases in Saccharomyces cerevisiae . J Biol Chem 276, 41710–41716. [DOI] [PubMed] [Google Scholar]
- 25. Athenstaedt K and Daum G (2000) 1‐Acyldihydroxyacetone‐phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J Biol Chem 275, 235–240. [DOI] [PubMed] [Google Scholar]
- 26. Benghezal M, Roubaty C, Veepuri V, Knudsen J and Conzelmann A (2007) SLC1 and SLC4 encode partially redundant acyl‐coenzyme A 1‐acylglycerol‐3‐phosphate O‐acyltransferases of budding yeast. J Biol Chem 282, 30845–30855. [DOI] [PubMed] [Google Scholar]
- 27. Ghosh AK, Ramakrishnan G and Rajasekharan R (2008) YLR099C (ICT1) encodes a soluble acyl‐CoA‐dependent lysophosphatidic acid acyltransferase responsible for enhanced phospholipid synthesis on organic solvent stress in Saccharomyces cerevisiae . J Biol Chem 283, 9768–9775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ayciriex S, Le Guedard M, Camougrand N, Velours G, Schoene M, Leon S, Wattelet‐Boyer V, Dupuy J‐W, Shevchenko A, Schmitter J‐M et al (2012) YPR139c/LOA1 encodes a novel lysophosphatidic acid acyltransferase associated with lipid droplets and involved in TAG homeostasis. Mol Biol Cell 23, 233–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pascual F and Carman GM (2013) Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim Biophys Acta 1831, 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tamura Y, Harada Y, Nishikawa SI, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K et al (2013) Tam41 is a CDP‐diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab 17, 709–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shen H, Heacock PN, Clancey CJ and Dowhan W (1996) The CDS1 gene encoding CDP‐diacylglycerol synthase In Saccharomyces cerevisiae is essential for cell growth. J Biol Chem 271, 789–795. [DOI] [PubMed] [Google Scholar]
- 32. Letts VA, Klig LS, Bae‐Lee M, Carman GM and Henry SA (1983) Isolation of the yeast structural gene for the membrane‐associated enzyme phosphatidylserine synthase. Proc Natl Acad Sci U S A 80, 7279–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nikawa J and Yamashita S (1984) Molecular cloning of the gene encoding CDPdiacylglycerol‐inositol 3‐phosphatidyl transferase in Saccharomyces cerevisiae . Eur J Biochem 143, 251–256. [DOI] [PubMed] [Google Scholar]
- 34. Chang SC, Heacock PN, Clancey CJ and Dowhan W (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycerophosphate synthase of Saccharomyces cerevisiae . J Biol Chem 273, 9829–9836. [DOI] [PubMed] [Google Scholar]
- 35. Osman C, Haag M, Wieland FT, Brügger B and Langer T (2010) A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J 29, 1976–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang S, Heacock PN, Mileykovskaya E, Voelker DR and Dowhan W (1998) Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae . J Biol Chem 273, 14933–14941. [DOI] [PubMed] [Google Scholar]
- 37. Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F and Daum G (1998) YDL142c encodes cardiolipin synthase (Clslp) and is non‐essential for aerobic growth of Saccharomyces cerevisiae . FEBS Lett 421, 15–18. [DOI] [PubMed] [Google Scholar]
- 38. Jiang F, Rizavi HS and Greenberg ML (1997) Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non‐fermentable carbon sources. Mol Microbiol 26, 481–491. [DOI] [PubMed] [Google Scholar]
- 39. Voelker DR (1997) Phosphatidylserine decarboxylase. Biochim Biophys Acta 1348, 236–244. [DOI] [PubMed] [Google Scholar]
- 40. Kodaki T and Yamashita S (1987) Yeast phosphatidylethanolamine methylation pathway ‐ cloning and characterization of 2 distinct methyltransferase genes. J Biol Chem 262, 15428–15435. [PubMed] [Google Scholar]
- 41. Gibellini F and Smith TK (2010) The Kennedy pathway‐de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62, 414–428. [DOI] [PubMed] [Google Scholar]
- 42. Huh W‐KK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS and O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- 43. Czabany T, Athenstaedt K and Daum G (2007) Synthesis, storage and degradation of neutral lipids in yeast. Biochim Biophys Acta 1771, 299–309. [DOI] [PubMed] [Google Scholar]
- 44. Han GS, O'Hara L, Carman GM and Siniossoglou S (2008) An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem 283, 20433–20442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ernst R, Ejsing CS and Antonny B (2016) Homeoviscous adaptation and the regulation of membrane lipids. J Mol Biol 428, 4776–4791. [DOI] [PubMed] [Google Scholar]
- 46. Martin CE, Oh CS and Jiang Y (2007) Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim Biophys Acta 1771, 271–285. [DOI] [PubMed] [Google Scholar]
- 47. Suutari M, Liukkonen K and Laakso S (1990) Temperature adaptation in yeasts: the role of fatty acids. J Gen Microbiol 136, 1469–1474. [DOI] [PubMed] [Google Scholar]
- 48. Okuyama H, Saito M, Joshi VC, Gunsberg S and Wakil SJ (1979) Regulation by temperature of the chain length of fatty acids in yeast. J Biol Chem 254, 12281–12284. [PubMed] [Google Scholar]
- 49. Hori T, Nakamura N and Okuyama H (1987) Possible involvement of acetyl coenzyme A carboxylase as well as fatty acid synthetase in the temperature‐controlled synthesis of fatty acids in Saccharomyces cerevisiae . J Biochem 101, 949–956. [DOI] [PubMed] [Google Scholar]
- 50. Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD and Tartakoff AM (1996) A yeast acetyl coenzyme A carboxylase mutant links very‐long‐chain fatty acid synthesis to the structure and function of the nuclear membrane‐pore complex. Mol Cell Biol 16, 7161–7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schneiter R, Guerra CE, Lampl M, Tatzer V, Zellnig G, Klein HL and Kohlwein SD (2000) A novel cold‐sensitive allele of the rate‐limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol Cell Biol 20, 2984–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hofbauer HF, Schopf FH, Schleifer H, Knittelfelder OL, Pieber B, Rechberger GN, Wolinski H, Gaspar ML, Kappe CO, Stadlmann J et al (2014) Regulation of gene expression through a transcriptional repressor that senses acyl‐chain length in membrane phospholipids. Dev Cell 29, 729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD and Jentsch S (2001) Activation of a membrane‐bound transcription factor by regulated ubiquitin/proteasome‐dependent processing. Chemtracts 14, 148–151. [DOI] [PubMed] [Google Scholar]
- 54. Rape M, Hoppe T, Gorr I, Kalocay M, Richly H and Jentsch S (2001) Mobilization of processed, membrane‐tethered SPT23 transcription factor by CDC48UFD1/NPL4, a ubiquitin‐selective chaperone. Cell 107, 667–677. [DOI] [PubMed] [Google Scholar]
- 55. Surma MA, Klose C, Peng D, Shales M, Mrejen C, Stefanko A, Braberg H, Gordon DE, Vorkel D, Ejsing CS et al (2013) A lipid E‐MAP identifies Ubx2 as a critical regulator of lipid saturation and lipid bilayer stress. Mol Cell 51, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Covino R, Ballweg S, Stordeur C, Michaelis JB, Puth K, Wernig F, Bahrami A, Ernst AM, Hummer G and Ernst R (2016) A eukaryotic sensor for membrane lipid saturation. Mol Cell 63, 49–59. [DOI] [PubMed] [Google Scholar]
- 57. Ballweg S and Ernst R (2017) Control of membrane fluidity: the OLE pathway in focus. Biol Chem 398, 215–228. [DOI] [PubMed] [Google Scholar]
- 58. De Smet CH, Vittone E, Scherer M, Houweling M, Liebisch G, Brouwers JF and de Kroon AIPM (2012) The yeast acyltransferase Sct1p regulates fatty acid desaturation by competing with the desaturase Ole1p. Mol Biol Cell 23, 1146–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shui G, Guan XL, Gopalakrishnan P, Xue Y, Goh JSY, Yang H and Wenk MR (2010) Characterization of substrate preference for Slc1p and Cst26p in Saccharomyces cerevisiae using lipidomic approaches and an LPAAT activity assay. PLoS One 5, e11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oelkers K and Pokhrel K (2016) Four acyltransferases uniquely contribute to phospholipid heterogeneity in Saccharomyces cerevisiae . Lipid Insights 9, 31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jain S, Stanford N, Bhagwat N, Seiler B, Costanzo M, Boone C and Oelkers P (2007) Identification of a novel lysophospholipid acyltransferase in Saccharomyces cerevisiae . J Biol Chem 282, 30562–30569. [DOI] [PubMed] [Google Scholar]
- 62. Riekhof WR, Wu J, Gijón MA, Zarini S, Murphy RC and Voelker DR (2007) Lysophosphatidylcholine metabolism in Saccharomyces cerevisiae: the role of P‐type ATPases in transport and a broad specificity acyltransferase in acylation. J Biol Chem 282, 36853–36861. [DOI] [PubMed] [Google Scholar]
- 63. Riekhof WR, Wu J, Jones JL and Voelker DR (2007) Identification and characterization of the major lysophosphatidylethanolamine acyltransferase in Saccharomyces cerevisiae . J Biol Chem 282, 28344–28352. [DOI] [PubMed] [Google Scholar]
- 64. Tamaki H, Shimada A, Ito Y, Ohya M, Takase J, Miyashita M, Miyagawa H, Nozaki H, Nakayama R and Kumagai H (2007) LPT1 encodes a membrane‐bound O‐acyltransferase involved in the acylation of lysophospholipids in the yeast Saccharomyces cerevisiae . J Biol Chem 282, 34288–34298. [DOI] [PubMed] [Google Scholar]
- 65. Sandager L, Gustavsson MH, Ståhl U, Dahlqvist A, Wiberg E, Banas A, Lenman M, Ronne H and Stymne S (2002) Storage lipid synthesis is non‐essential in yeast. J Biol Chem 277, 6478–6482. [DOI] [PubMed] [Google Scholar]
- 66. Black PN and DiRusso CC (2007) Yeast acyl‐CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim Biophys Acta 1771, 286–298. [DOI] [PubMed] [Google Scholar]
- 67. Mora G, Scharnewski M and Fulda M (2012) Neutral lipid metabolism influences phospholipid synthesis and deacylation in Saccharomyces cerevisiae . PLoS One 7, e49269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Horvath SE and Daum G (2013) Lipids of mitochondria. Prog Lipid Res 52, 590–614. [DOI] [PubMed] [Google Scholar]
- 69. Bürgermeister M, Birner‐Grünberger R, Heyn M and Daum G (2004) Contribution of different biosynthetic pathways to species selectivity of aminoglycerophospholipids assembled into mitochondrial membranes of the yeast Saccharomyces cerevisiae . Biochim Biophys Acta 1686, 148–160. [DOI] [PubMed] [Google Scholar]
- 70. Schneiter R, Brügger B, Sandhoff R, Zellnig G, Leber A, Lampl M, Athenstaedt K, Hrastnik C, Eder S, Daum G et al (1999) Electrospray ionization tandem mass spectrometry (ESI‐MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain‐based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol 146, 741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Heikinheimo L and Somerharju P (2002) Translocation of phosphatidylthreonine and ‐serine to mitochondria diminishes exponentially with increasing molecular hydrophobicity. Traffic 3, 367–377. [DOI] [PubMed] [Google Scholar]
- 72. Boumann HA, Damen MJA, Versluis C, Heck AJR, De Kruijff B and De Kroon AIPM (2003) The two biosynthetic routes leading to phosphatidylcholine in yeast produce different sets of molecular species. Evidence for lipid remodeling. Biochemistry 42, 3054–3059. [DOI] [PubMed] [Google Scholar]
- 73. Renne MF, Bao X, De Smet CH and de Kroon AIPM (2015) Lipid acyl chain remodeling in yeast. Lipid Insights 2015, 33–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Fisher E, Almaguer C, Holic R, Griac P and Patton‐Vogt J (2005) Glycerophosphocholine‐dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae . J Biol Chem 280, 36110–36117. [DOI] [PubMed] [Google Scholar]
- 75. Fernández‐Murray JP and McMaster CR (2005) Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J Biol Chem 280, 38290–38296. [DOI] [PubMed] [Google Scholar]
- 76. Stålberg K, Neal AC, Ronne H and Ståhl U (2008) Identification of a novel GPCAT activity and a new pathway for phosphatidylcholine biosynthesis in S. cerevisiae . J Lipid Res 49, 1794–1806. [DOI] [PubMed] [Google Scholar]
- 77. Głab B, Beganovic M, Anaokar S, Hao MS, Rasmusson AG, Patton‐Vogt J, Banaś A, Stymne S and Lager I (2016) Cloning of glycerophosphocholine acyltransferase (GPCAT) from fungi and plants: a novel enzyme in phosphatidylcholine synthesis. J Biol Chem 291, 25066–25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Waksman M, Eli Y, Liscovitch M and Gerst JE (1996) Identification and characterization of a gene encoding phospholipase D activity in yeast. J Biol Chem 271, 2361–2364. [DOI] [PubMed] [Google Scholar]
- 79. Wagner S and Paltauf F (1994) Generation of glycerophospholipid molecular species in the yeast Saccharomyces cerevisiae. Fatty acid pattern of phospholipid classes and selective acyl turnover at sn‐1 and sn‐2 positions. Yeast 10, 1429–1437. [DOI] [PubMed] [Google Scholar]
- 80. Kishino H, Eguchi H, Takagi K, Horiuchi H, Fukuda R and Ohta A (2014) Acyl‐chain remodeling of dioctanoyl‐phosphatidylcholine in Saccharomyces cerevisiae mutant defective in de novo and salvage phosphatidylcholine synthesis. Biochem Biophys Res Commun 445, 289–293. [DOI] [PubMed] [Google Scholar]
- 81. De Smet CH, Cox R, Brouwers JF and De Kroon AIPM (2013) Yeast cells accumulate excess endogenous palmitate in phosphatidylcholine by acyl chain remodeling involving the phospholipase B Plb1p. Biochim Biophys Acta 1831, 1167–1176. [DOI] [PubMed] [Google Scholar]
- 82. Tanaka K, Fukuda R, Ono Y, Eguchi H, Nagasawa S, Nakatani Y, Watanabe H, Nakanishi H, Taguchi R and Ohta A (2008) Incorporation and remodeling of extracellular phosphatidylcholine with short acyl residues in Saccharomyces cerevisiae . Biochim Biophys Acta 1781, 391–399. [DOI] [PubMed] [Google Scholar]
- 83. Los D and Murata N (2004) Membrane fluidity and its role in the perception of environmental signals. Biochim Biophys Acta 1666, 142–157. [DOI] [PubMed] [Google Scholar]
- 84. Lewis RNAH, Mak N and McElhaney RN (1987) A differential scanning calorimetric study of the thermotropic phase behavior of model membranes composed of phosphatidylcholines containing linear saturated fatty acyl chains. Biochemistry 26, 6118–6126. [DOI] [PubMed] [Google Scholar]
- 85. Van Dijck PWM, De Kruijff B, Van Deenen LLM, De Gier J and Demel RA (1976) The preference of cholesterol for phosphatidylcholine in mixed phosphatidylcholine‐phosphatidylethanolamine bilayers. Biochim Biophys Acta 455, 576–587. [DOI] [PubMed] [Google Scholar]
- 86. Boggs JM (1987) Lipid intermolecular hydrogen bonding: influence on structural organization and membrane function. Biochim Biophys Acta 906, 353–404. [DOI] [PubMed] [Google Scholar]
- 87. Dawaliby R, Trubbia C, Delporte C, Noyon C, Ruysschaert JM, Van Antwerpen P and Govaerts C (2016) Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J Biol Chem 291, 3658–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liebisch G, Vizcaíno JA, Köfeler H, Trötzmüller M, Griffiths WJ, Schmitz G, Spener F and Wakelam MJO (2013) Shorthand notation for lipid structures derived from mass spectrometry. J Lipid Res 54, 1523–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Gennis RB (1989) Biomembranes. Springer, New York, NY. [Google Scholar]
- 90. Koynova RD, Boyanov AI and Tenchov BG (1985) On the phase diagram of an L‐dipalmitoylphosphatidylcholine/cholesterol mixture. FEBS Lett 187, 65–68. [Google Scholar]
- 91. Sharpe HJ, Stevens TJ and Munro S (2010) A comprehensive comparison of transmembrane domains reveals organelle‐specific properties. Cell 142, 158–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Munro S (1998) Localization of proteins to the Golgi apparatus. Trends Cell Biol 8, 11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Cybulski LE, Ballering J, Moussatova A, Inda ME, Vazquez DB, Wassenaar TA, de Mendoza D, Tieleman DP and Killian JA (2015) Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc Natl Acad Sci U S A 112, 6353–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Cybulski LE, Martín M, Mansilla MC, Fernández A and De Mendoza D (2010) Membrane thickness cue for cold sensing in a bacterium. Curr Biol 20, 1539–1544. [DOI] [PubMed] [Google Scholar]
- 95. Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hänelt I, Hummer G and Ernst R (2017) Activation of the unfolded protein response by lipid bilayer stress. Mol Cell 67 673–684.e8. [DOI] [PubMed] [Google Scholar]
- 96. Cullis PR and de Kruijff B (1979) Lipid polymorphism and the functional roles of lipids in biological membranes. Biochim Biophys Acta 559, 399–420. [DOI] [PubMed] [Google Scholar]
- 97. Gruner SM (1985) Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proc Natl Acad Sci U S A 82, 3665–3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Koynova R and Caffrey M (1994) Phases and phase‐transitions of the hydrated phosphatidylethanolamines. Chem Phys Lipids 69, 1–34. [DOI] [PubMed] [Google Scholar]
- 99. Van den Brink‐van der Laan E, Antoinette Killian J and De Kruijff B (2004) Nonbilayer lipids affect peripheral and integral membrane proteins via changes in the lateral pressure profile. Biochim Biophys Acta 1666, 275–288. [DOI] [PubMed] [Google Scholar]
- 100. Boumann HA, Gubbens J, Koorengevel MC, Oh C‐S, Martin CE, Heck AJR, Patton‐Vogt J, Henry SA, de Kruijff B and de Kroon AIPM (2006) Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains: evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol Biol Cell 17, 1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Cantor RS (1997) The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry 36, 2339–2344. [DOI] [PubMed] [Google Scholar]
- 102. Marsh D (1996) Lateral pressure in membranes. Biochim Biophys Acta 1286, 183–223. [DOI] [PubMed] [Google Scholar]
- 103. Van den Brink‐van der Laan E, Chupin V, Killian JA and De Kruijff B (2004) Stability of KcsA tetramer depends on membrane lateral pressure. Biochemistry 43, 4240–4250. [DOI] [PubMed] [Google Scholar]
- 104. Cantor RS (1999) Lipid composition and the lateral pressure profile in bilayers. Biophys J 76, 2625–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Cantor RS (1999) The influence of membrane lateral pressures on simple geometric models of protein conformational equilibria. Chem Phys Lipids 101, 45–56. [DOI] [PubMed] [Google Scholar]
- 106. Van den Brink‐van der Laan E, Chupin V, Killian JA and De Kruijff B (2004) Small alcohols destabilize the KcsA tetramer via their effect on the membrane lateral pressure. Biochemistry 43, 5937–5942. [DOI] [PubMed] [Google Scholar]
- 107. Drin G and Antonny B (2010) Amphipathic helices and membrane curvature. FEBS Lett 584, 1840–1847. [DOI] [PubMed] [Google Scholar]
- 108. Cornell RB (2016) Membrane lipid compositional sensing by the inducible amphipathic helix of CCT. Biochim Biophys Acta 1861, 847–861. [DOI] [PubMed] [Google Scholar]
- 109. Karanasios E, Han G‐S, Xu Z, Carman GM and Siniossoglou S (2010) A phosphorylation‐regulated amphipathic helix controls the membrane translocation and function of the yeast phosphatidate phosphatase. Proc Natl Acad Sci U S A 107, 17539–17544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Bodenmiller B, Wanka S, Kraft C, Urban J, Campbell D, Pedrioli PG, Gerrits B, Picotti P, Lam H, Vitek O et al (2010) Phosphoproteomic analysis reveals interconnected system‐wide responses to perturbations of kinases and phosphatases in yeast. Sci Signal 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Hancock SE, Poad BLJ, Batarseh A, Abbott SK and Mitchell TW (2017) Advances and unresolved challenges in the structural characterization of isomeric lipids. Anal Biochem 524, 45–55. [DOI] [PubMed] [Google Scholar]
- 112. Davis PJ, Fleming BD, Coolbear KP and Keough KMW (1981) Gel to liquid‐crystalline transition temperatures of water dispersions of two pairs of positional isomers of unsaturated mixed‐acid phosphatidylcholines. Biochemistry 20, 3633–3636. [DOI] [PubMed] [Google Scholar]
- 113. Barton PG and Gunstone FD (1975) Hydrocarbon chain packing and molecular motion in phospholipid bilayers formed from unsaturated lecithins. Synthesis and properties of sixteen positional isomers of 1,2‐dioctadecenoyl‐sn‐glycero‐3‐phosphorylcholine. J Biol Chem 250, 4470–4476. [PubMed] [Google Scholar]
- 114. Lewis RN and McElhaney RN (1993) Calorimetric and spectroscopic studies of the polymorphic phase behavior of a homologous series of n‐saturated 1,2‐diacyl phosphatidylethanolamines. Biophys J 64, 1081–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Wang ZQ, Lin HN, Li S and Huang CH (1994) Calorimetric studies and molecular mechanics simulation of monounsaturated phosphatidylethanolamine bilayers. J Biol Chem 269, 23491–23499. [PubMed] [Google Scholar]
- 116. Wang Z, Lin H, Li S and Huang C (1995) Phase transition behavior and molecular structures of monounsaturated phosphatidylcholines. J Biol Chem 270, 2014–2023. [DOI] [PubMed] [Google Scholar]
- 117. Seddon JM, Cevc G and Marsh D (1983) Calorimetric studies of the gel‐fluid (L.beta.‐L.alpha.) and lamellar‐inverted hexagonal (L.alpha.‐HII) phase transitions in dialkyl‐ and diacylphosphatidylethanolamines. Biochemistry 22, 1280–1289. [DOI] [PubMed] [Google Scholar]
- 118. Epand RM (1985) High sensitivity differential scanning calorimetry of the bilayer to hexagonal phase transitions of diacylphosphatidylethanolamines. Chem Phys Lipids 36, 387–393. [Google Scholar]
- 119. Epand RM (1990) Hydrogen bonding and the thermotropic transitions of phosphatidylethanolamines. Chem Phys Lipids 52, 227–230. [Google Scholar]
- 120. Marsh D (2013) Handbook of lipid bilayers, 2nd edn CRC Press, Boca Raton, FL. [Google Scholar]


