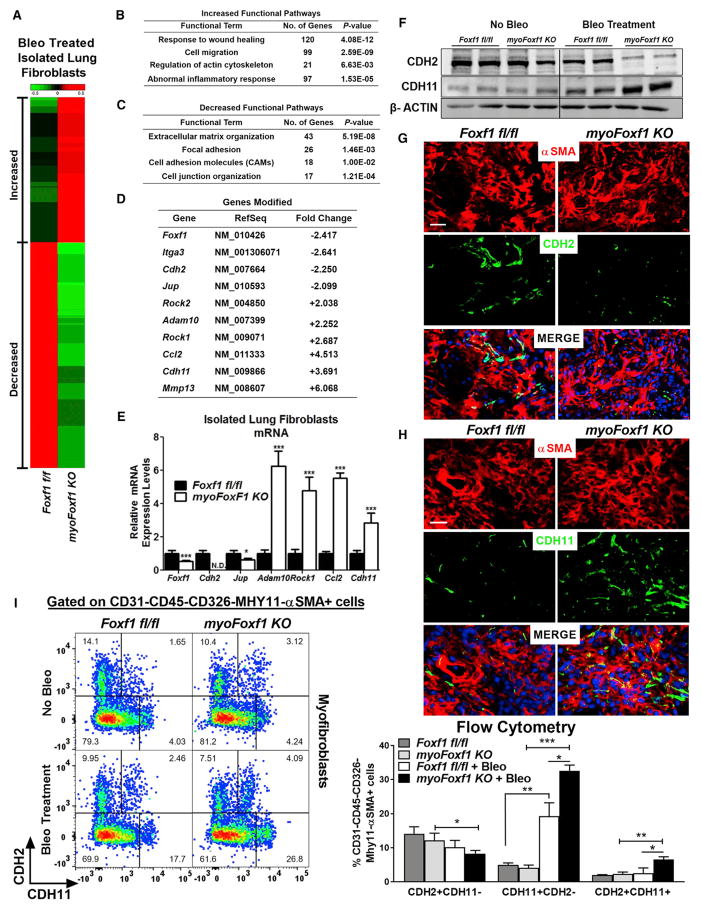
Figure 5. FOXF1 Regulates Genes Essential for Myofibroblast Differentiation and Functions.
(A) Differentially expressed genes in bleomycin-treated myoFoxf1 KO lungs were identified using RNA-seq analysis. Heatmap of RNA-seq results showing 2,809 coding genes significantly upregulated (red) or downregulated (green) between isolated fibroblasts from bleomycin-treated Foxf1fl/fl and those from myoFoxf1 KO lungs. Samples from three mice per genotype were pooled for RNA-seq analysis.
(B and C) Increased (B) and decreased (C) functional pathways influenced by Foxf1 deletion were identified using the ToppGene Suite (https://toppgene.cchmc.org/). The statistical significance of each functional pathway was presented using negative log2 transformation of the p value.
(D) Fold changes in mRNA for several genes from the RNA-seq mapped in (A).
(E) qRT-PCR validation of several genes found to be differentially expressed by RNA-seq analysis. n = 7–8 mice per genotype. See also Figure S5.
(F) FOXF1 regulates CDH2-CDH11 cadherin switching in mouse lung fibroblasts. Representative images of western blot analysis of CDH2 and CDH11 levels in isolated lung fibroblast fractions from Foxf1fl/fl and myoFoxf1 KO lungs. n = 7–8 mice per group.
(G and H) Immunofluorescence costaining of (G) αSMA and CDH2 or (H) CDH11 in fibrotic lesions of Foxf1fl/fl and myoFoxf1 KO lungs. Representative images of the staining are shown for n = 9–11 mice. Scale bars indicate 20 μm.
(I) FACS shows an increase in the percentage of CDH11+CDH2− and CDH2+CDH11+ myofibroblasts (CD45−CD326−CD31−MHY11−αSMA+) in bleomycin-treated myoFoxf1 KO lungs. Representative dot plots and quantification of the percentage of CDH2+CDH11−, CDH11+CDH2−, and CDH2+CDH11+ myofibroblasts are shown. n = 4 mice/group.
Data presented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. See also Figure S6.