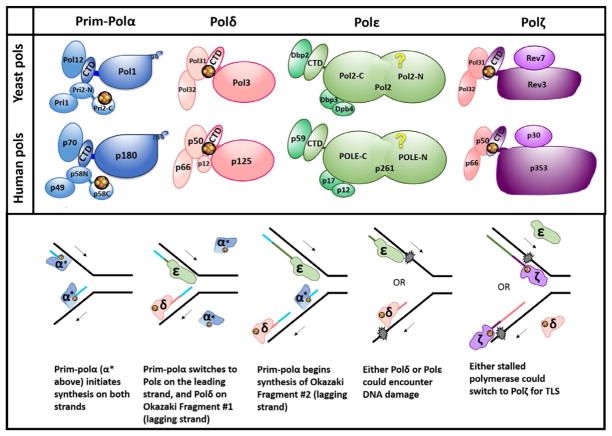
Fig. 1.
B-family DNA polymerases at the replication fork. (A) Subunit structure of yeast and human B-family DNA polymerases. Groups of yellow and orange spheres represent [4Fe–4S] clusters. Only those clusters supported by multiple studies (see Burgers & Kunkel, 2017) are depicted here; others have been proposed but have not been verified yet. These include clusters in CTDs of Polα and Polε. Names of subunits for human Pols corresponding to their molecular mass are shown; the names of the subunits are as follows (from larger to smaller): Polα: POLA1, POLA2, PRIM2, PRIM1; Polδ: POLD1–POLD4; Polε: POLE, POLE2, POLE3, POLE4; Polζ: REV3L, hREV7 (or MAD2L2), POLD3, POLD4. See detailed nomenclature in (Baranovskiy, Gu, et al., 2017). (B) DNA replication under normal conditions and in the presence of DNA damage. Iron–sulfur clusters are shown as in (A). Arrows indicate the direction of DNA synthesis on the respective strand (leading strand top, lagging strand bottom). Newly synthesized DNA is color coded to match the Pol that synthesized that stretch. Blue represents both the primer laid by primase and the DNA laid by Polα. Pol colors match panel (A). For concision, the word “pol” has been omitted to leave only the greek letters to denote Pols, and α* refers to pol α (dark blue portion) plus primase (light blue portion). Recent evidence has suggested that Polδ may also play a role in leading strand synthesis under certain conditions (Burgers & Kunkel, 2017), but this is not shown here.