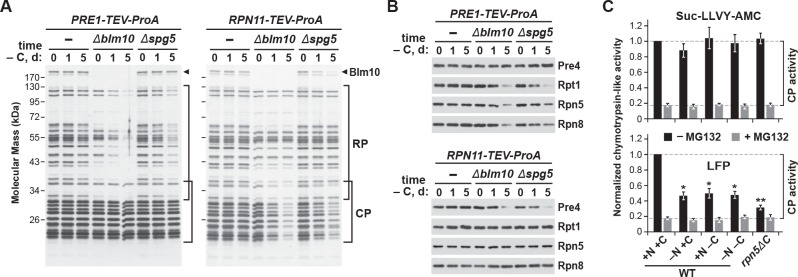
Figure 6. The CP and RP dissociate from each other upon carbon starvation and are separately delivered into PSGs.
(A and B) Yeast proteasomes selectively lose the RP or CP sub-complexes when purified from Δblm10 and Δspg5 cells via the CP or RP, respectively, upon growth on –C medium. PRE1-TEV-ProA or RPN11-TEV-ProA cells with or without the Δblm10 or Δspg5 mutations were switched from nutrient-rich (+N +C) medium to –C medium for the indicated times before affinity purification of proteasomes based on their ProA tags in the presence of ATP. The enriched proteasomes were subjected to SDS-PAGE followed by either staining for total protein with silver (panel A) or by immunoblotting with antibodies specific to subunits of the CP (Pre4) or RP (Rpt1, Rpn5 or Rpn8; panel B). In panel A, the distributions of the core CP and RP subunits are indicated by the brackets, and the position of Blm10 is indicated by the arrowheads. (C) Proteasome CPs remain active under conditions that promote PSG formation, but are less associated with the RP. Cells were grown on +N +C medium and then switched to media lacking either nitrogen (–N), carbon (–C), or both (–N –C) for 1 day. Total protein extracts were then assayed for CP peptidase activity using either Suc-LLVY-amc or Mca-AKVYPYPME-(Dpa)Dnp-amide (LFP) substrates that monitor total CP activity or RP-dependent CP activity, respectively. Black and grey bars represent the mean chymotrypsin-like activity (±SD) in the absence and presence of MG132, respectively, from three independent biological replicates, each comprised of three technical replicates.