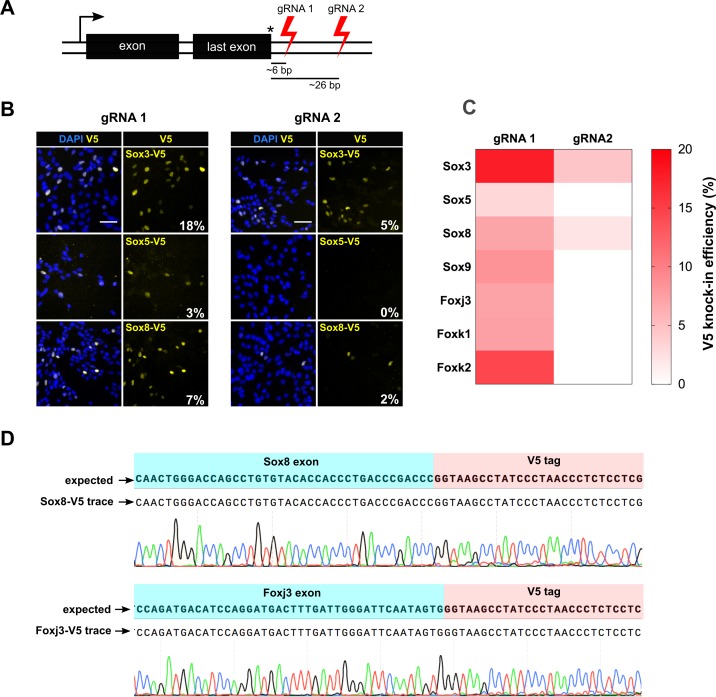
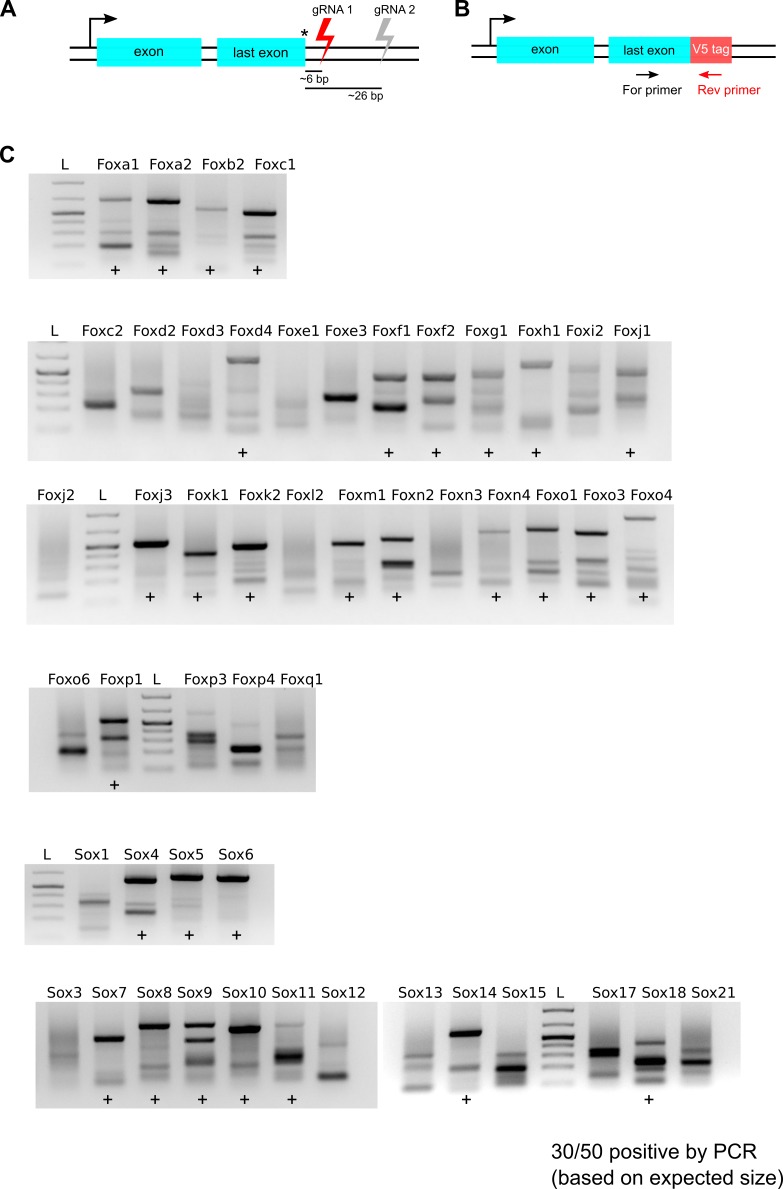
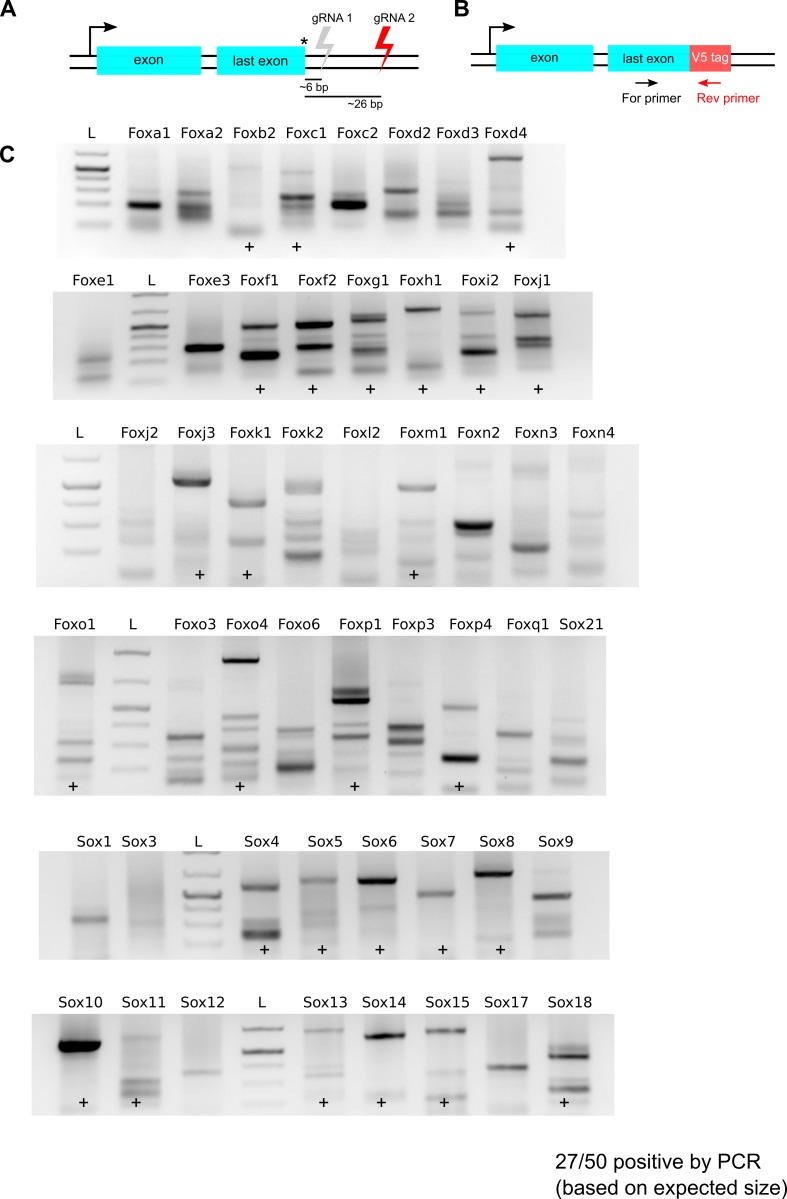
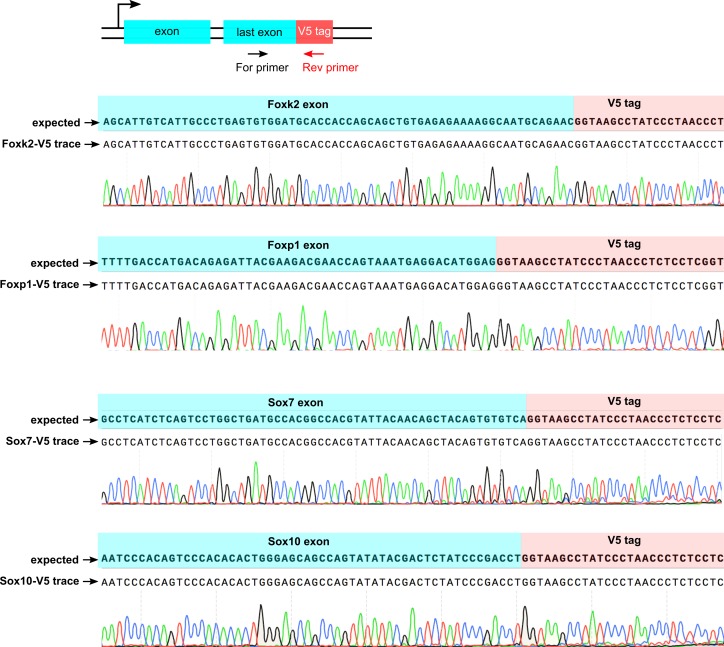
Figure 4. csRNP cutting proximal to insertion site is more efficient and allows knock-in at non-expressed genes.
(A) Schematic showing the relative position of guide RNAs in the 3’ UTR. The first set of gRNA (gRNA1) cuts proximal to the stop codon, the second set (gRNA 2) cuts distal. Average distance of cut site from the insertion site is shown for both sets of gRNAs against 50 TFs. (B) Representative V5 ICC images of Sox3, Sox5, and Sox8 V5 knock-in bulk populations obtained with gRNA 1 (left panels) and gRNA 2 (right panels) are shown; % V5 knock-in efficiency for each experiment is shown at the right bottom of V5 panels (numbers in white). (C) Heatmap representation of V5 knock-in efficiency obtained with gRNA1 and gRNA2 for different genes, color code is shown on the right. (D) Example of sequencing traces using the bulk populations of V5-tagged cells for Sox8 and Foxj3. Alignment with the expected TF-V5 chimeric sequence is shown.