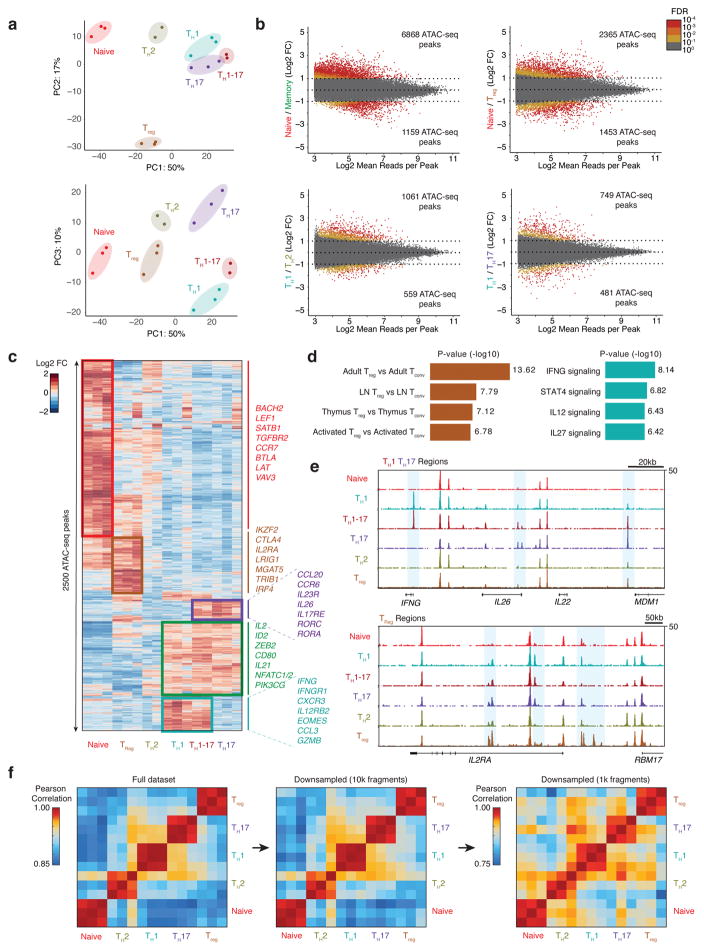
Figure 3. Epigenomic landscape of ensemble human CD4+ T cell subtypes.
(a) PCA of ensemble ATAC-seq profiles from CD4+ T cell subtypes using the top 2500 variable ATAC-seq peaks (as defined by variance rank of log2 variance-stabilized read counts; n=3, 3 independent experiments). Percentages indicate percent of variance explained by each PC. (b) Differential ATAC-seq peaks for the indicated T cell subtypes. Memory T cell signatures reflect the average accessibility in TH1, TH2, TH17, and TH1-17 cells. (c) Heat map showing clusters for top 2500 varying ATAC-seq peaks. Colors indicate log2 fold-change of reads in each peak compared to the mean across all T cell types. (d) Left, MSigDB Immunologic Signatures of Treg-specific ATAC-seq peaks as obtained from GREAT analysis. Right, MSigDB Pathway Signatures of TH1-specific ATAC-seq peaks as obtained from GREAT analysis (Binomial test, n=3, 3 independent experiments). (e) Ensemble ATAC-seq data genome tracks for the indicated T cell subtypes. Highlighted regions show cell type-specific ATAC-seq peaks. (f) Pearson correlation of PC scores of ensemble ATAC-seq profiles and of ensemble ATAC-seq profiles after downsampling each profile to 10,000 or 1,000 fragments. Downsampling was performed by randomly selecting 10,000 or 1,000 nuclear fragments in each ensemble ATAC-seq .bam file. Heat maps demonstrate that CD4+ T cell subtype profiles can be distinguished from one another using the full dataset or profiles with a fraction of the fragments, as expected in single-cell libraries (16 ATAC-seq profiles obtained from 3 independent experiments).