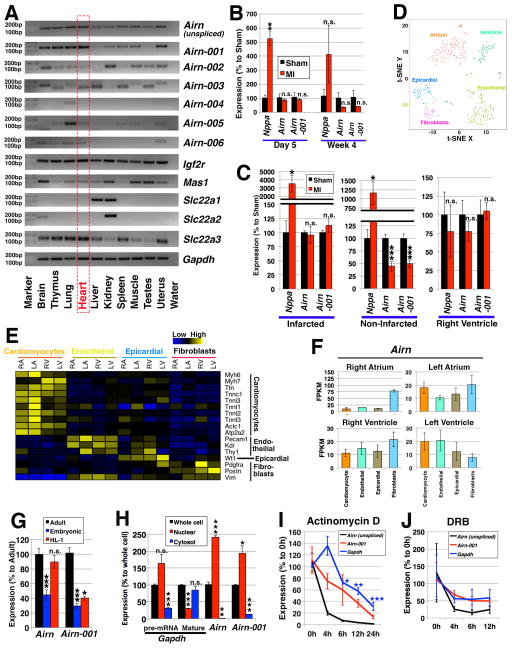
Figure 1. Airn and its isoforms.
(A) Expression profiling of Airn and its isoforms in mouse tissues. (B, C) Expression in myocardial infarction (MI). (B) The time-course data of 5 days and 4 weeks post surgery comparing “Sham” control and “MI” mice. n=3 mice for sham (day 5) and n=4 mice for all the other conditions. The expressions were normalized to those of sham samples using Gapdh as internal control. (C) Expression in different regions of hearts 4 weeks after the operation. One heart was divided into 3 regions: “Infarcted”; “Non-Infarcted” (i.e., border and remote regions); and “Right Ventricle”. n=5 mice. The expressions were normalized to those of sham samples using Rn18s as internal control. (D-F) Expression profiling of single-cell RNA-seq data of E10.5 mouse hearts. (D) 5 clusters of cells were defined: cardiomyocytes from atrium or ventricle, endothelial cells, epicardial cells, and fibroblasts. (E) Expressions of marker genes. (F) Expression patterns of the Airn gene. (G) Expression of unspliced Airn (“Airn”) and Airn-001 in “adult” cardiomyocytes compared to those of “embryonic” cardiomyocytes and “HL-1” cells. n=4 samples for adult cardiomyocytes and n=3 samples for others. (H) Subcellular localization in HL-1 cells. As controls, primer pairs for Gapdh of its pre-mRNA (targeting the intron between its exon 2 and 3) and mature RNA (targeting exons 4 and 5) were used. Statistical calculations were made against the expressions in the whole cell. n=3. (I, J) Stability of mRNAs upon the treatment with (I) actinomycin D at 4, 6, 12 and 24 hours; and (J) DRB at 4, 6 and 12 hours. n=3. The expressions were normalized using those of Rn18s. *, ** and *** represent p<0.05, 0.01 and 0.005, respectively. “n.s.” represents “not statistically significant”.