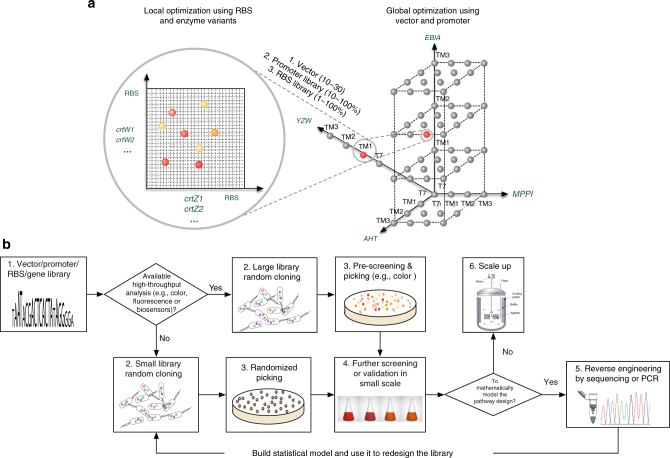
Fig. 4.
Illustration of MHP and its workflow. a In MHP, pathway engineering proceeds either sequentially or simultaneously with global optimization (promoters) and local refinement (5′UTRS or RBSs and enzyme variants). Globally, each promoter regulates the transcription of each operon/module with multiple genes (such as module AHT, MPPI, EBIA and YZW). Locally, various RBSs and enzymes facilitate fine tuning of the relative activities within one specific module (such as crtW and crtZ gene in YZW module). b The MHP workflow. First, libraries of promoters and RBSs were established and characterized either experimentally or mathematically. It was followed by a decision node that depends on the availability of high-throughput analytical methods of products (colorimetry or fluorescence). If so, the subsequent randomized cloning proceeded with a large combinatorial library (1010–1015) and the resulting strain library were pre-screened by colony color or fluorescence. Otherwise, the cloning continued with a small combinatorial library (102–103), followed by randomized picking and analyzed by low/medium throughput analysis (gas chromatography and liquid chromatography, etc). Next, performances of the strain were validated in liquid culture and desired phenotypes were further reversely engineered. The data could be then used to build statistical models and to refine new library to further improve phenotypes. Lastly, the optimized strain will be scaled up in bioreactors