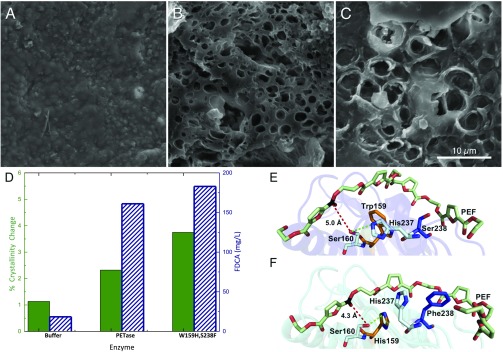
Fig. 4.
Comparison of PETase and the engineered enzyme S238F/W159H with PEF. (A) Buffer-only control of PEF coupon. (B) PEF coupon after incubation with wild-type PETase. (C) PEF coupon after incubation with the PETase double mutant, S238F/W159H. All SEM images were taken after 96 h of incubation at a PETase loading of 50 nM (pH 7.2) in phosphate buffer or a buffer-only control. (Scale bar: A–C, 10 μm.) (D) Percent crystallinity change (green, solid bar) and reaction product concentration (FDCA, blue diagonal lines) after incubation with buffer, wild-type PETase, and the S238F/W159H double mutant. (E) Predicted binding conformations of wild-type PETase from docking simulations demonstrate that PEF is accommodated in an optimum position for the interaction of the carbon (black) with the nucleophilic hydroxyl group of Ser160, at a distance of 5.0 Å (red dash). His237 is positioned within 3.7 Å of the Ser160 hydroxyl (green dash). Residues Trp159 (orange) and Ser238 (blue) line the active-site channel. (F) In contrast, the double mutant S238F/W159H significantly alters the architecture of the catalytic site for PEF binding. Residue His237 rotates away from Ser160, and instead forms an aromatic interaction with PEF chain at 5.1 Å. Surprisingly, the mutated His159 becomes an alternative productive H-bond partner at 3.2 Å. Similar to interactions with PET, Phe238 also provides additional hydrophobic interactions to an adjacent furan ring of the extended PEF polymer, creating a more intimate binding mode with the cleft, with a parallel displaced aromatic interaction at 5.2 Å.