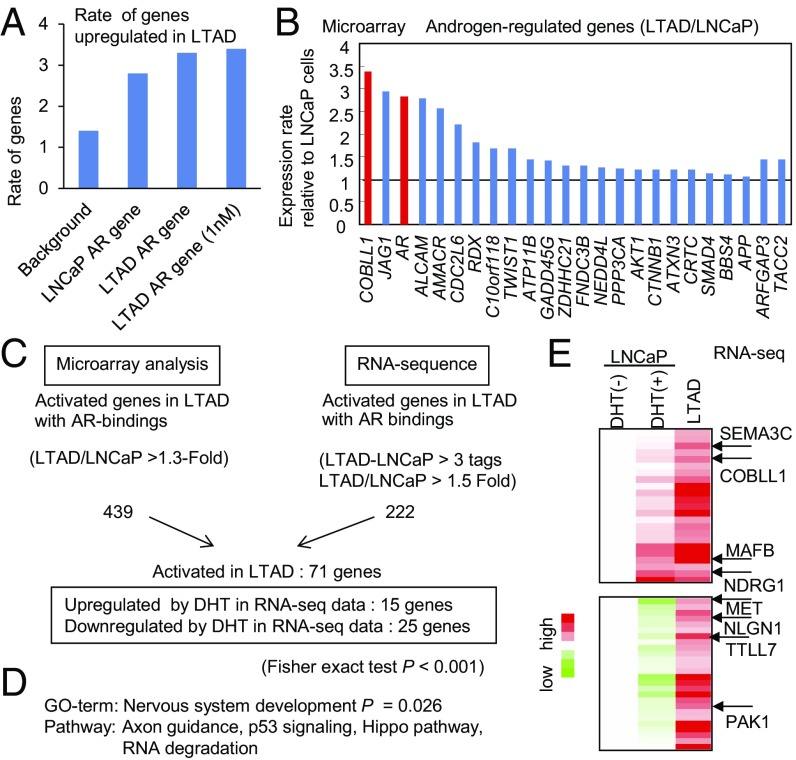
Fig. 1.
Identification of AR-induced genes up-regulated in the absence of androgen in LTAD CRPC model cells by combining microarray, RNA-seq, and ChIP-seq analysis. (A) AR-binding genes are up-regulated significantly in LTAD CRPC model cells. We identified AR-binding genes in LNCaP and LTAD cells in the presence of DHT (10 or 1 nM) by using ChIP-seq data. Up-regulated genes in LTAD cells were found by microarray analysis. (B) COBLL1 is highly up-regulated in LTAD CRPC model cells compared with parental LNCaP cells. By microarray analysis, we analyzed the expression level of AR-induced genes and AR in LTAD cells (without androgen treatment). Fold changes over the expression level in LNCaP cells are shown. (C) Summary of up-regulated AR-binding genes in LTAD CRPC model cells by RNA-seq and microarray analysis. (D) Pathway and GO analysis of AR-binding genes up-regulated in LTAD CRPC model cells. (E) The top 50 genes up-regulated in LTAD CRPC model cells. The results of RNA-seq are summarized in the heat map. Representative genes associated with neuronal function are indicated.