Significance
G protein-coupled receptors (GPCRs) continue to be the pharmaceutical target of over one-third of FDA-approved drugs, yet our understanding of GPCR function has been overwhelmingly limited to the study of intracellular effectors. It is emerging that GPCRs also associate with a variety of extracellular proteins, yet the functional consequences of these interactions have been ill defined for the lack of robust approaches to measure them. In this study, we develop a unique assay platform and provide an example of transcellular modulation of GPCR properties by association with a synaptic adhesion molecule.
Keywords: GPCR, synapse, cell adhesion, extracellular interactions, transsynaptic
Abstract
Functional characterization of the GPCR interactome has been focused predominantly on intracellular interactions, yet GPCRs are increasingly found in complex with extracellular proteins. Extracellular leucine-rich repeat fibronectin type III domain containing 1 (ELFN1) was recently reported to physically anchor mGluR6 and mGluR7 across retinal and hippocampal synapses, respectively; however, the consequence of transsynaptic interactions on properties and pharmacology of these receptors are unknown. In the current study, we explore the effects of ELFN1 on mGluR signaling and pharmacology. First, we established the binding specificity of ELFN1 and found it to be recruited selectively to all group III mGluRs (mGluR4, mGluR6, mGluR7, and mGluR8), but not other mGluR species. Using site-directed mutagenesis we mapped binding determinants of this interaction to two distinct sites on the ELFN1 ectodomain. To evaluate functional aspects of the interaction, we developed a transcellular signaling assay in reconstituted HEK293 cells which monitors changes in mGluR activity in one cell following its exposure to separate ELFN1-containing cells. Using this platform, we found that ELFN1 acts as an allosteric modulator of class III mGluR activity in suppressing cAMP accumulation: altering both agonist-induced and constitutive receptor activity. Using bioluminescence resonance energy transfer-based real-time kinetic assays, we established that ELFN1 alters the ability of mGluRs to activate G proteins. Our findings demonstrate that core properties of class III mGluRs can be altered via extracellular interactions with ELFN1 which serves as a transsynaptic allosteric modulator for these receptors. Furthermore, our unique assay platform opens avenues for exploring transcellular/transsynaptic pharmacology of other GPCR transcomplexes.
The meticulous coordination of neurotransmission involving a myriad of mediators requires precise spatial organization and matching of neurotransmitter release machinery with cognate receptors across compatible synapses (1–3). This structural organization must be further coupled to the temporally synchronous release of many neurotransmitters at diverse synapses endowed with distinct properties (4). The importance of intricate spatial and temporal coordination of neurotransmission is emphasized by correlations between dysfunction in these elements of neural circuitry and the manifestation of neuropsychiatric disease (5–7). However, how the structure–functional organization of transsynaptic matching is achieved and modulated remains poorly understood.
G protein-coupled receptors (GPCRs) constitute the largest family of neurotransmitter receptors and represent one of the most common targets of current pharmaceutical treatment strategies (8). Functional characterization of GPCRs and the GPCR interactome has focused predominantly on intracellular transducers and regulators (9–11). However, it is becoming increasingly clear that GPCRs also associate with extracellular partners in trans, and that these interactions play an important role in their biology. Perhaps the best known example of transsynaptic complexes involving GPCRs is provided by the adhesion GPCRs latrophilins (LPHNs) whose interactions in extracellular space with teneurin-2/4 (12–14), fibronectin leucine-rich transmembrane protein (FLRT) 2/3 (15), and neurexin1–3 (16) play important roles in synapse formation and function. However, examples of such interactions are scarce and our knowledge of the functional implications, as well as the extent in which different GPCRs engage in transcellular complexes, is very limited.
Metabotropic glutamate receptors (mGluRs) represent a prominent family of GPCRs that participate in the neuromodulation of synaptic transmission (17, 18). mGluRs are subtyped into three groups largely based on sequence homology. Prototypically, group I mGluRs (mGluR1/5) are Gαq-coupled and localized predominantly postsynaptically in the brain where they modulate neuronal excitability in response to excitatory inputs. Conversely, group II (mGluR2/3) and group III mGluRs (mGluR4/6/7/8) are Gαi/o-coupled and predominantly localized presynaptically in the brain where they participate in the autoregulation of neurotransmitter release (17). Although this spatial organization is fairly conserved, there are exceptions at distinct synapses, most notably with group III mGluR6 in retinal synapses that functions as a postsynaptic neurotransmitter receptor integral for the ON bipolar response to light stimulation (19). Of the remaining group III mGluRs, mGluR4, -7, and -8 can be found in a plethora of different brain regions where they have been implicated in the manifestation and/or treatment of a myriad of neurological and neuropsychiatric diseases, including Parkinson’s disease, neuropathic pain, epilepsy, anxiety, depression, and autism-like symptoms, among others (20–26). Therefore, a thorough understanding of group III mGluR function and pharmacology is prerequisite for the development of new therapeutic strategies for the treatment of an extensive array of neurological and neuropsychiatric diseases.
Recently, two members of the group III mGluRs were found to interact with extracellular leucine-rich repeat and fibronectin type III domain-containing 1 (ELFN1): an understudied protein that has been implicated in neurological diseases such as epilepsy, attention deficit hyperactivity, and autism spectrum disorders (27, 28). ELFN1 belongs to a large family of leucine-rich repeat (LRR) neuronal adhesion proteins that have been suggested to play an integral role in synapse formation and differentiation via the coordination of both pre- and postsynaptic machineries (2, 3, 29, 30). In the hippocampal synapse, ELFN1 was first reported as a postsynaptic retrograde regulator of presynaptic release probability acting on fine-tuning properties of select neural circuits (31). Subsequent studies determined postsynaptic ELFN1 to be an essential transsynaptic scaffolding protein for the recruitment of presynaptic metabotropic glutamate receptor 7 (mGluR7): known for its role as an autoreceptor capable of altering presynaptic release (28). In the retinal synapse, ELFN1 is targeted to the presynaptic terminal of rod photoreceptors via an association with neurotransmitter release machinery (32, 33). There, it transsynaptically binds to postsynaptic receptor mGluR6 and this interaction plays an essential role in the retention of mGluR6 at the synapse, synaptic transmission, and the physical wiring of rods (32, 33). Taken together, transsynaptic ELFN1-mGluR6/7 complexes have been demonstrated to be spatially organized across distinct synapses where they contribute to key synaptic properties. However, our understanding of functional aspects of ELFN1-mGluR interaction, and transsynaptic complexes involving GPCRs in general, at the mechanistic level remains in its infancy: creating a void in our knowledge of GPCR function and pharmacology.
To advance our understanding of transsynaptic GPCR complexes, in the current study we have developed a unique assay platform to study the functional consequence of transcellular interactions on GPCR signaling and pharmacology. We document that ELFN1 is a global extracellular adaptor for all group III mGluRs that critically changes the pharmacological properties of these receptors by promoting constitutive activity and constraining agonist-mediated signaling. We suggest these actions fine tune ELFN1-positive synapses by endowing synapses with distinct functional properties while stabilizing the optimal spatial localization of group III mGluRs.
Results
ELFN1 Interacts with Multiple mGluRs with Selectivity for All Members of Group III.
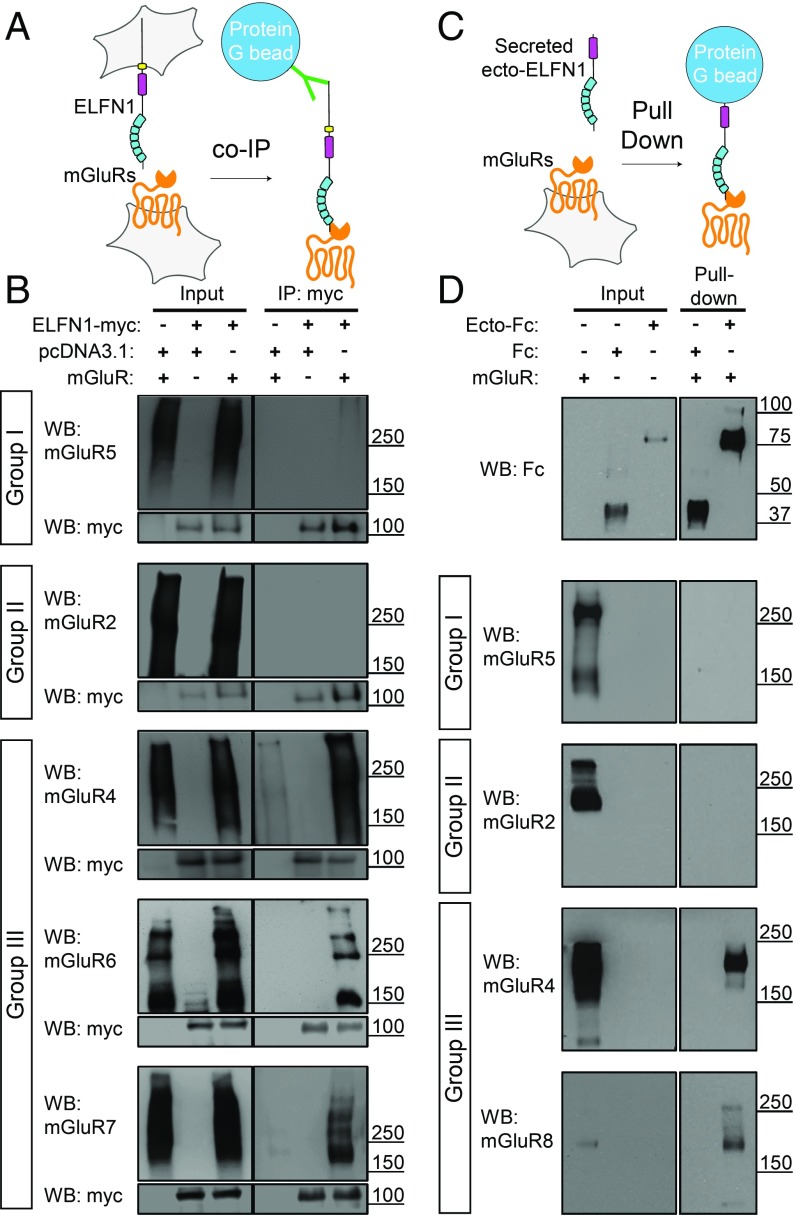
Given that ELFN1 has been reported to interact with more than one mGluR, we began by defining its interaction selectivity across members of the mGluR family from groups I–III. Following transfection into HEK293 cells, ELFN1 was immunoprecipitated with specific antibodies and incubated with lysates of separate cells expressing mGluRs (Fig. 1A). We found that ELFN1 effectively coimmunoprecipitated with mGluR4, mGluR6, and mGluR7, which belong to group III, but not mGluR5 or mGluR2, which belong to groups I and II, respectively (Fig. 1B). To determine whether ELFN1 ectodomain mediates these interactions, the extracellular portion of ELFN1 sequence was fused with an Fc tag, thereby directing its secretion. Following its capture from the culture media by protein G beads, it was incubated with separate cell lysates expressing various mGluRs (Fig. 1C). In agreement with full-length ELFN1 coimmunoprecipitations, these pull-down assays revealed retention of group III mGluR4 and mGluR8, but not group I mGluR5 or group II mGluR2 by ELFN1 ectodomain, demonstrating sufficiency for the interaction (Fig. 1D). Together, these experiments identify mGluR4 and mGluR8 as unique interacting partners of ELFN1 and establish ELFN1 as a selective binding partner of all group III mGluRs.
Fig. 1.
ELFN1 interacts with multiple mGluRs with selectivity for all members of group III. (A) Schematic representation of coimmunoprecipitation methodology whereby ELFN1 in one cell population was immunoprecipitated and screened for coimmunoprecipitation of mGluRs from another cell population’s lysate. (B) Western blots of cell lysates and immunoprecipitations with myc antibody conjugated to protein G beads. (C) Schematic representation of protein G pull-down methods whereby secreted Fc-conjugated ELFN1 ectodomain was incubated with various mGluR-expressing cell lysates and protein G beads. (D) Western blots with inputs and pull-downs with ELFN1 Ecto-Fc conjugated to protein G beads.
Multiple Structural Elements Within ELFN1 Ectodomain Mediate Its Interaction With mGluRs.
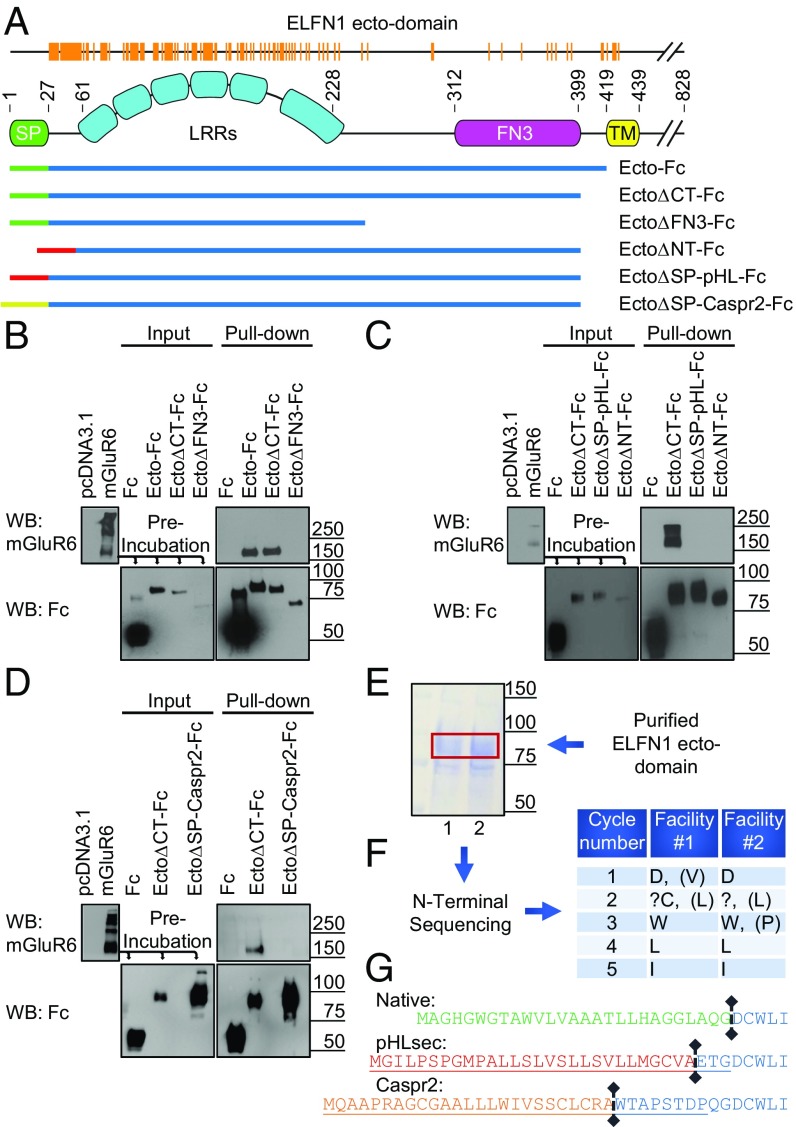
ELFN1 contains several structural domains within the extracellular region (Fig. 2A), including multiple leucine-rich repeats (LRRs) and a fibronectin type III (FN3) domain. To investigate the structural requirements of ELFN1 for mGluR interactions, we performed site-directed mutagenesis on the ELFN1 ectodomain. After performing a multiple sequence alignment across 165 species of ELFN1 proteins, we identified the invariant residues within the ectodomain that show 100% conservation (Fig. 2A, orange) and engineered several deletional Fc-tagged constructs omitting the following conserved regions: the ectodomain carboxyl terminus (EctoΔCT), FN3 domain (EctoΔFN3), the amino terminal (EctoΔNT), and signal peptide cleavage sites (EctoΔSP-pHL and EctoΔSP-Caspr2) (Fig. 2A). Following incubation of culture media from various ectodomain-expressing HEK293 cells with lysates containing mGluR6, we observed that EctoΔCT retained mGluR6 as well as intact ectodomain. However, EctoΔFN3 was unable to pull down mGluR6 (Fig. 2B), suggesting the FN3 domain of ELFN1 is important for the interaction with mGluRs.
Fig. 2.
Multiple structural elements within ELFN1 ectodomain mediate its interaction with mGluRs. (A) Multiple sequence alignment of 165 species of ELFN1 ectodomain reveal 100% conserved residues (orange) aligned with domain topology. cDNA constructs developed to dissect binding determinants are aligned to these structures below. (B) Evaluation of mutants with deletions in carboxyl terminal regions of ELFN1 ectodomain (EctoΔCT-Fc and EctoΔFN3-Fc) for the interaction with mGluR6 in pull-down assays. (C) Evaluation of mutants with deletions in amino terminal regions of ELFN1 ectodomain (EctoΔNT-Fc and EctoΔSP-pHL-Fc) for interaction with mGluR6. (D) The effect of changing the signal peptide (EctoΔSP-Caspr2-Fc) on the interaction of ELFN1 with mGluR6. (E) Analysis of Fc-tagged Ecto constructs carrying native signal peptide purified with protein G beads from culture media by gel electrophoresis followed by Coomassie staining. (F) Results of two separate facilities performing amino terminal sequencing by Edman degradation of purified ELFN1 ectodomain proteins. (G) Experimentally determined signal peptide cleavage site and predictions by Phobius software for native sequence and constructs carrying pHLsec (EctoΔSP-pHL) and Caspr2 (EctoΔSP-Caspr2-Fc) signal peptides. Predicted cleavage sites are denoted by the vertical lines and appended signal peptides sequences are underlined.
We next probed the contribution of the highly conserved ELFN1 distal amino terminus to binding. For these experiments, we cloned the ELFN1 ectodomain construct into a pHLsec vector for optimized expression of extracellular ectodomains (34), which incidentally entailed changing the signal peptide. We found that deletion of a small part of the distal N-terminal sequence (ELFN1ΔNT) completely abolished ELFN1 interactions with mGluR6 (Fig. 2C). Surprisingly, we further observed that even ELFN1 with presumably intact N-terminal sequence expressed from the pHLsec vector (EctoΔSP-pHL) was also completely deficient in mGluR6 binding, suggesting that the identity of the signal peptide may be important for binding. To test this hypothesis, we supplied ELFN1 with yet another signal peptide (derived from protein Caspr2), while maintaining the N-terminal sequence intact (Fig. 2A). Curiously, EctoΔSP-Caspr2, like EctoΔSP-pHL, was completely deficient in mGluR6 binding (Fig. 2D).
This led us to experimentally determine the identity of the ELFN1 N-terminal sequence and cleavage site directed by the native signal peptide to determine the required posttranslational processing necessary for mGluR interactions. The Fc-tagged ectodomain of ELFN1 construct (ELFN1-Ecto-Fc) carrying native signal peptide was purified with protein G beads from culture media following secretion, yielding protein of the expected molecular weight as detected by Coomassie staining (Fig. 2E). N-terminal sequencing by Edman degradation of the corresponding band at two separate facilities independently identified the first five amino acids to match the DCWLI sequence in ELFN1 (Fig. 2F), which perfectly matched the prediction by Phobius software (phobius.sbc.su.se/) (Fig. 2G), thus defining the cleavage site and pinpointing aspartic acid to be the first amino acid of the mature ELFN1 capable of interacting with mGluRs. We next used the same software to predict cleavage sites and identities of N-terminal amino acids in nonbinding constructs with transplanted Caspr2 and pHLsec signal peptides. This analysis revealed that predicted cleavage sites of these signal peptides alter the sequence upstream of the aspartic acid, adding 10 and 3 amino acids to the amino terminus, respectively (Fig. 2G). This suggests that the exact sequence identity of the extreme N terminus of ELFN1 is essential for its interaction with mGluRs, and neither additions nor deletions are tolerated. Together, our mapping experiments identify that ELFN1 interaction with mGluRs are dependent on two determinants: the ELFN1 distal amino terminus initiated with aspartic acid and the FN3 domain.
ELFN1 Modulates Pharmacological Properties of Group III mGluRs in Trans.
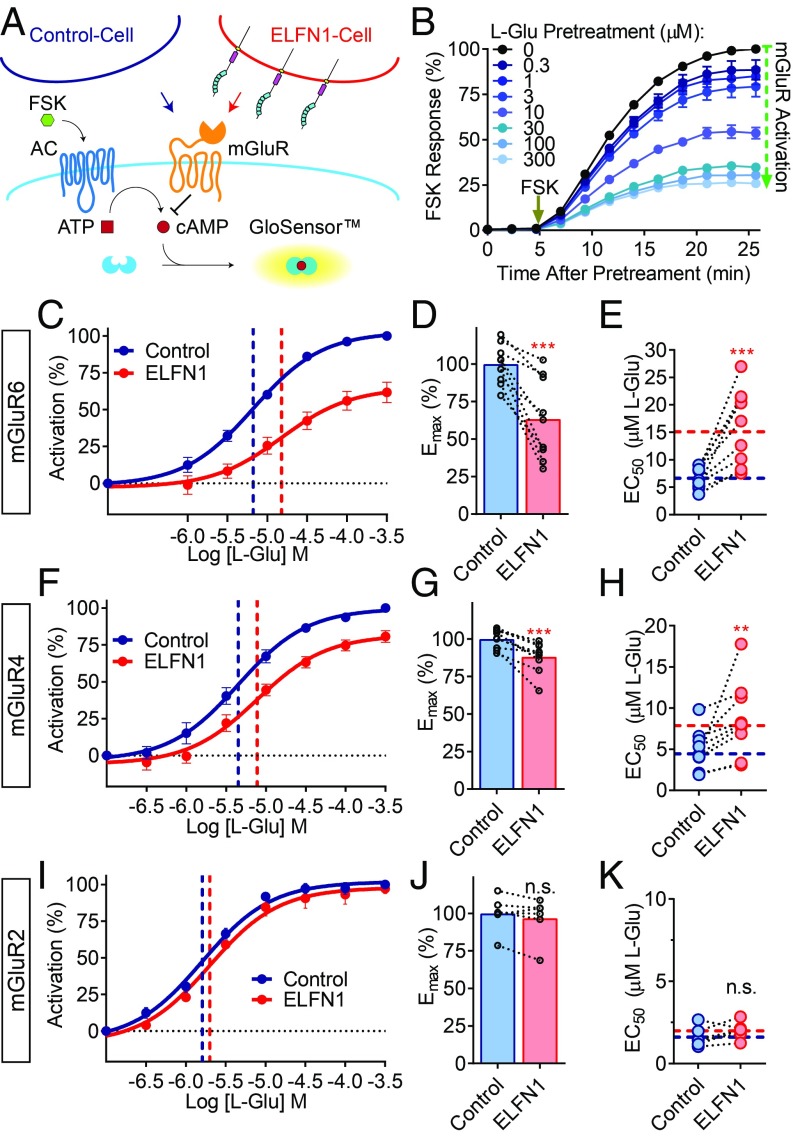
To address the functional consequences of ELFN1 interaction with group III mGluRs, we developed a unique assay platform for probing effects of transcellular protein–protein interactions on GPCR signaling (Fig. 3A). The assay takes advantage of measuring the suppression of cAMP by mGluR expressed in one set of cells in the presence of another set of cells providing ELFN1 as a binding partner in trans. We utilized the -22F cAMP pGloSensor to monitor forskolin-induced increases in cAMP levels in HEK293 cells expressing mGluRs activated by various levels of agonists. Dose dependence studies show that, similar to previous reports (35), stimulation of mGluR with increasing concentrations of l-glutamic acid led to progressive reduction in forskolin-mediated cAMP response, which we used as a measure of mGluR activation (Fig. 3B). By design, cAMP response originated only from the mGluR-expressing cells, and identical cell populations were then exposed to either mock-transfected control cells or ELFN1-expressing cells to reconstitute transcellular interactions (Fig. 3A).
Fig. 3.
ELFN1 modulates pharmacological properties of group III mGluRs in trans. (A) Schematic representation of transcellular GPCR signaling assay utilizing -22F cAMP pGloSensor in mGluR-expressing cells exposed to control or ELFN1-expressing cells that lack biosensor. (B) Luminescence reading of cells pretreated with various concentrations of l-glutamic acid for 5 min and then stimulated with 1 µM forskolin, with greater luminescence attributed to greater levels of cAMP. (C) Dose–response curves were plotted for the same mGluR6- and biosensor-expressing cell population, differing only by exposure to either control or ELFN1-expressing cells. Identical cultures indicated by dotted lines. (D) Quantification of maximal efficacy (Emax) and (E) half maximal effective concentration (EC50) for l-glutamic acid. (F) Dose–response curves for mGluR4- and biosensor-expressing cells exposed to control or ELFN1-expressing cells. (G) Quantification of Emax and (H) EC50 for l-glutamic acid. (I) Dose–response curves for mGluR2- and biosensor-expressing cells exposed to control or ELFN1-expressing cells. (J) Quantification of Emax and (K) EC50 for l-glutamic acid.
We observed that exposure to ELFN1-expressing cells correlated with a very modest but reproducible elevation of the baseline mGluR6 activity in the absence of agonist (SI Appendix, Fig. S1A). Exposure to ELFN1 significantly decreased the maximum efficacy (Emax) (Fig. 3 C and D) and increased the half maximal effective concentration (EC50) of mGluR6 activation by l-glutamic acid (Fig. 3 C and E) as demonstrated by the rightward shift in the concentration–response curve (Fig. 3C), resembling the effects of negative allosteric modulators (24). We observed a similar decrease in the Emax (SI Appendix, Fig. S2 A and B) and an increase in the EC50 (SI Appendix, Fig. S2 A and C) when stimulating mGluR6 with the group III-selective orthosteric agonist l-2-amino-4-phosphonobutyric acid (l-AP4) in the presence of transcellularly supplied ELFN1.
We further tested the effects of ELFN1 on mGluR4, another member of group III, which we found to also be interacting with ELFN1, and observed very similar effects. Exposure to ELFN1 transcellularly similarly correlated with very modest increases in basal activity of mGluR4 (SI Appendix, Fig. S1B) and also caused a decrease in the Emax (Fig. 3 F and G) and an elevation of the EC50 for both l-glutamic acid (Fig. 3 F and H) and l-AP4 (SI Appendix, Fig. S2 D–F). To test whether this effect can be specifically attributable to ELFN1–mGluR interactions, we performed a control experiment with a group II receptor mGluR2, which does not bind ELFN1. Like mGluR4 and mGluR6, mGluR2 couples to Gαi/o and causes substantial inhibition of cAMP production (Fig. 3I). However, we found that exposure to the same level of transcellular ELFN1 had no effect on basal activity (SI Appendix, Fig. S1C), Emax (Fig. 3 I and J), or EC50 (Fig. 3 I and K). Additionally, mutant transmembrane ELFN1 constructs ELFN1ΔFN3 and Caspr2-ELFN1 deficient in mGluR binding had no effect on mGluR6 Emax or EC50 (SI Appendix, Fig. S3). Together, these data indicate that the transcellular interaction of group III mGluRs with its extracellular binding partner ELFN1 modulates key pharmacological properties of these GPCRs.
Reduction of Group III mGluRs Efficacy by ELFN1 Is Not a Consequence of Altered Desensitization.
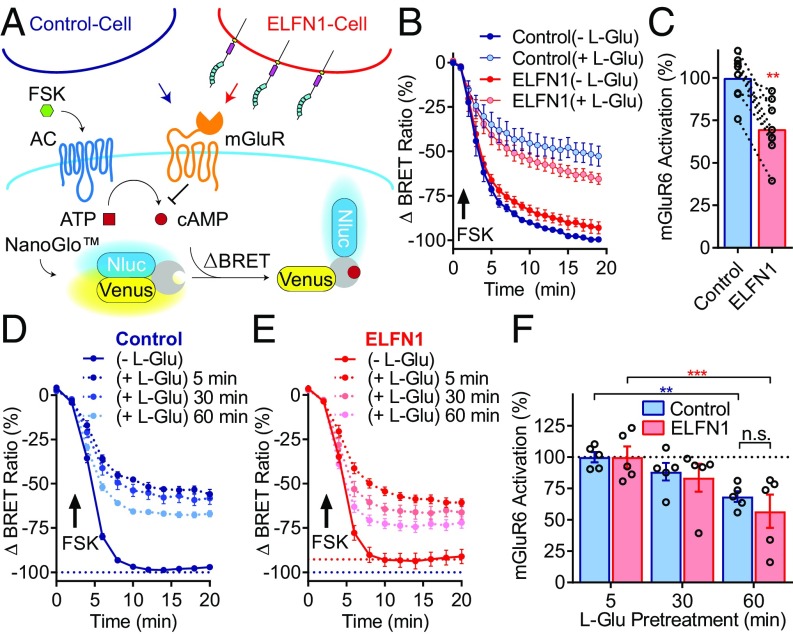
To probe whether ELFN1 modulation of group III mGluR efficacy was a consequence of altered receptor activity over time, we evaluated acute receptor desensitization. Due to limitations with longitudinal desensitization experiments with the luciferase-based pGloSensor platform, we modified our transcellular assays to monitor changes in cAMP by the genetically encoded bioluminescence resonance energy transfer (BRET)-based cAMP sensor that permits longitudinal evaluation of signaling in real time (Fig. 4A). This sensor reports increased cAMP accumulation as a decrease in BRET ratio and the activity of group III mGluRs was assessed by their inhibition of forskolin-mediated cAMP accumulation, which suppressed BRET signal change. In congruence with measurements using the -22F cAMP pGloSensor platform, transcellular ELFN1 interactions constrained the maximal efficacy of l-glutamic acid, as evidenced by a lower extent of mGluR6-mediated suppression of forskolin-driven cAMP production (Fig. 4 B and C). Once established, we began longitudinal measurements following stimulation of mGluR6 with 300 µM l-glutamic acid for 5, 30, or 60 min in the presence of either control cells (Fig. 4D) or cells expressing ELFN1 (Fig. 4E). Conditions reported previously to facilitate group III mGluR desensitization were successful in producing moderate but significant desensitization of mGluR6 (Fig. 4F) (36). However, we observed no significant alterations in the rate of mGluR desensitization in the absence or presence of ELFN1, with equal loss of efficacy after 30 or 60 min of l-glutamic acid stimulation in both conditions (Fig. 4F). Furthermore, biotinylation experiments confirm mGluR6 was found at similar levels at the membrane in both control and ELFN1 conditions (SI Appendix, Fig. S4). Thus, these data suggest that modulation of group III mGluR efficacy by ELFN1 is not simply a consequence of altered receptor desensitization or membrane localization.
Fig. 4.
Reduction of group III mGluRs efficacy by ELFN1 is not a consequence of altered desensitization. (A) Schematic representation of transcellular GPCR signaling assay involving coculture of control cells or ELFN1-expressing cells with mGluR6-expressing cells coexpressing BRET-based Nluc-EPAC-VV cAMP biosensor. (B) Change in BRET ratio measurements over time of cells pretreated with buffer or 300 µM l-glutamic acid for 5 min and then stimulated with 1 µM forskolin, with increased cAMP equating to decreased BRET ratio. mGluR activation was quantified as a decrease in forskolin response. (C) Quantification of mGluR6 activation following coculture with control or ELFN1-expressing cells. (D) Change in BRET ratio readings following adaptation of assay to accommodate longitudinal prestimulations with 300 µM l-glutamic acid. The solid control line corresponds to no prestimulation/baseline in the presence of control cells, followed by 5, 30, or 60 min of l-glutamic acid prestimulation, with 5 min providing the maximum (Max) response. (E) Change in BRET ratio under the same parameters in parallel in the presence of ELFN1-expressing cells. (F) Quantification of mGluR6 activation normalizing for Max response (5 min).
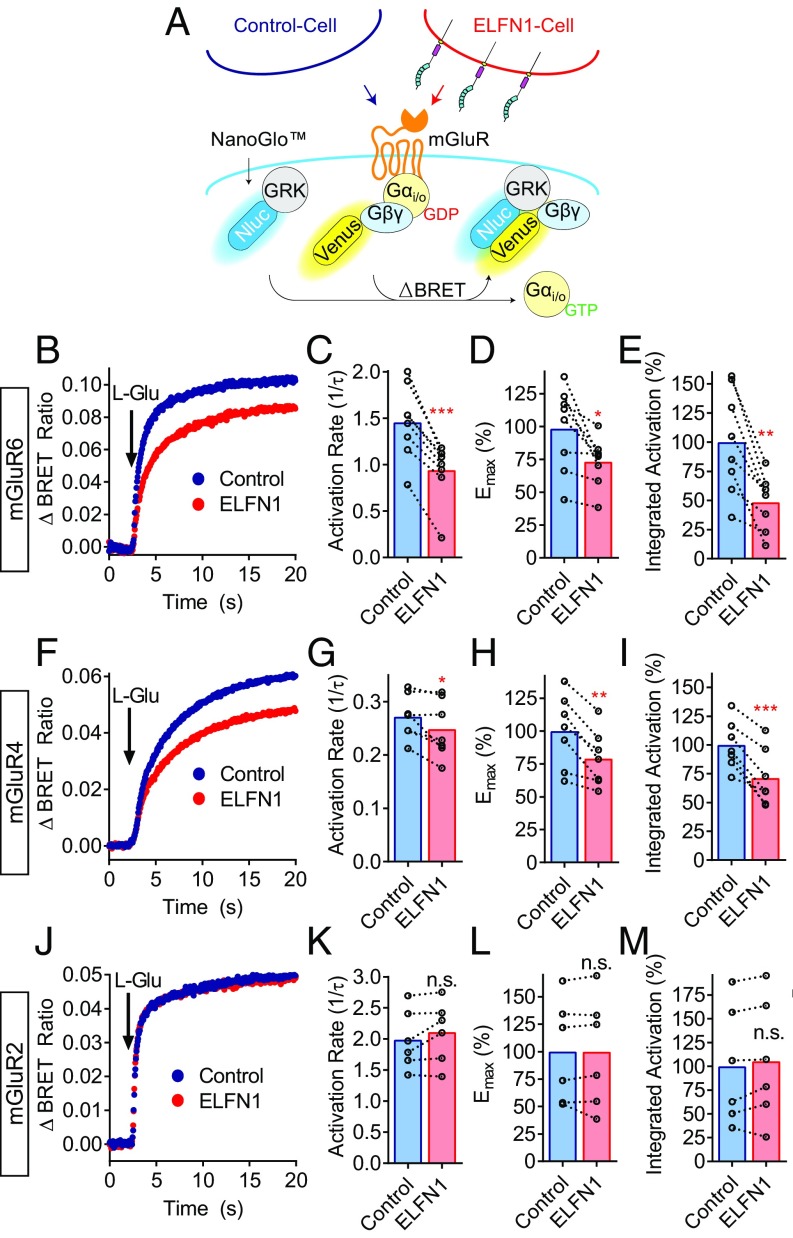
Transcellular Interaction with ELFN1 Alters G Protein-Coupling Efficiency of Group III mGluRs.
To dissect the mechanism of group III mGluR modulation of secondary messenger accumulation by transcellular ELFN1, we directly evaluated the ability of mGluRs to activate G proteins by adapting a highly sensitive BRET-based real-time kinetic assay (37) to a transcellular format (Fig. 5A). This assay monitors guanine nucleotide exchange factor (GEF) activity of a GPCR that triggers release of the Gβγ subunits upon agonist stimulation on a millisecond scale, while evaluating both maximal efficacy of response and the catalytic rate. We first studied ELFN1 impact on the ability of mGluR6 to activate its physiological substrate Gαo. We found that transcellular interaction with ELFN1 substantially decelerated the rate of Gαo activation (1/τ) in response to mGluR6 activation by l-glutamic acid (Fig. 5 B and C). Furthermore, the maximal extent of Gαo mobilization was diminished as evidenced by reduced maximum response amplitude (Emax) (Fig. 5 B and D). Similar negative modulation of agonist-mediated Gαo coupling by ELFN1 was observed with mGluR4 (Fig. 5 F–I). We next studied the activity of mGluR6 on Gαi, which likely was responsible for driving changes in cAMP in our initial experiments. Again, ELFN1 also reduced both activation rate and maximal amplitudes of Gαi activation by mGluR6, leading to a similar effect on net G protein activation, indicating that transcellular ELFN1 can alter both Gαo and Gαi activation (SI Appendix, Fig. S5). Notably, our control experiments with group II mGluR2, which does not bind ELFN1, showed no effect on its ability to activate Gαo, yielding identical efficacies (Fig. 5 J and K) and activation rates (Fig. 5 J and L), thereby demonstrating that observed effects were indeed driven by the interaction of group III mGluRs with ELFN1.
Fig. 5.
Transcellular interactions with ELFN1 alter G protein-coupling efficiency of group III mGluRs. (A) Schematic representation of transcellular GPCR signaling assay utilizing real-time kinetic Gαi/o coupling biosensors in mGluR-expressing cells cocultured with control or ELFN1-expressing cells. (B) Change in BRET ratio readings comparing coculture of control or ELFN1-expressing cells on average mGluR6 activation of Gαo, where increased BRET ratio corresponds to increased Gαo activation. (C) Quantification of mGluR6-mediated Gαo activation rate, (D) efficacy, and (E) integrated activation constant via l-glutamic acid. Identical cultures are indicated by dotted lines. (F) Change in BRET ratio readings comparing coculture of control or ELFN1-expressing cells on average mGluR4 activation of Gαo. (G) Quantification of mGluR4-mediated Gαo activation rate, (H) efficacy, and (I) integrated activation constant via l-glutamic acid. (J) Change in BRET ratio readings comparing coculture of control or ELFN1-expressing cells on average mGluR2 activation of Gαo. (K) Quantification of mGluR2-mediated Gαo activation rate, (L) efficacy, and (M) integrated activation constant via l-glutamic acid.
To evaluate the net G protein response, we calculated an integrative activation constant that combines effects of G protein-coupling rate and efficacy (Emax/τ) to better approximate downstream signaling consequences of transcellular ELFN1 interactions. ELFN1 reduced the integrated activation constant of both mGluR6 (Fig. 5E) and mGluR4 (Fig. 5I), but not mGluR2 (Fig. 5M), in good agreement with effects seen when measuring cAMP dynamics. Thus, we conclude that the mechanism for ELFN1’s transcellular regulation of group III mGluR downstream signaling involves allosteric modulation of receptor properties that lead to altered G protein activation.
Discussion
The intricate mechanisms guiding synaptic connectivity and differentiation of synaptic properties have increasingly been focused on extracellular neuronal adhesion molecules (2, 3, 29, 30, 38); however, the recent discovery of transsynaptic GPCR complexes provides an added complexity (12–16, 28, 32). The seminal transsynaptic GPCR complexes have been predominantly described for the adhesion GPCR family of LPHNs in transcomplex with teneurin-2/4 (12–14), FLRT2/3 (15), or neurexin1–3 (16). It is now evident from our study and others that the entirety of group III mGluRs can form transcomplexes with ELFN1 (28, 32), suggesting the large ectodomains of group III mGluRs can share some conserved signaling logic with organization of adhesion GPCRs like LPHNs.
Following identification of the extracellular transsynaptic interactions, one critical question pertains to their functional role which has been completely undefined. Addressing such a question is fundamental for our understanding of GPCR signaling mechanisms. However, it requires the development of assay platforms focused beyond defining mechanisms of intracellular modulators of GPCR signaling by traditional approaches. One strategy to achieve this goal was purification and administration of soluble ectodomain peptide fragments, which was successfully employed to define effects of teneurin-2–mediated induction of presynaptic Ca2+ release via LPHN1 (12). This strategy takes advantage of the inherent proteolytic cleavage and extracellular release of teneurin-2 ectodomain (14). However, it was subsequently demonstrated that soluble neurexin ectodomain was biologically ineffective and failed to drive synapse formation, an effect that was only observed when the neuronal adhesion molecule was membrane tethered (39). With these caveats in mind, we developed a unique assay platform for defining transcellular interactions involving GPCR signaling in living cells. We hope that such a platform will be useful for defining functional effects emanating from interactions of full-length GPCRs with native transmembrane adhesion molecules across intact cells, thereby recapitulating a more natural structural environment and constraints of these transcomplexes.
Using this platform, we provide a unique example of transcellular modulation of a GPCR’s pharmacological properties whereby ELFN1 selectively regulates group III mGluR-mediated signaling. Notably, transcellular interactions with ELFN1 may stabilize these receptors in a conformation amenable for constitutive activity as suggested by very modest heightening of basal signaling for mGluR6 and mGluR4. Upon stimulation with orthosteric agonists, ELFN1 constrains the efficacy and potency of the response, thereby acting in the capacity of an allosteric modulator. These observations are mirrored across multiple biosensors and are not simply a consequence of altered mGluR desensitization or membrane expression, but rather a direct modulation of conformational state that leads to G protein activation. We propose that this modulation of group III mGluRs may contribute to the ability of ELFN1 to fine tune synaptic properties and alter presynaptic release (31). In this model, postsynaptic ELFN1 could act as both a transsynaptic scaffold (28, 32) and a retrograde allosteric modulator of presynaptic group III mGluR autoreceptors to control presynaptic release probabilities and differentiate synaptic properties.
The recent observations for transcomplexing of neuronal adhesion proteins with GPCRs, and the present demonstrations that such interactions are functionally important, open an enormous void in our understanding of GPCR function in synaptic modulation. It is possible that extracellular interactions of many GPCRs, particularly those with large extracellular domains like adhesion and class C GPCRs, could be a common mechanism for regulating their signaling in native systems. It is foreseeable that analogous complexes could also be observed at other cell–cell junctions. The current study provides a unique framework for dissecting the consequence of identified transcomplexes on GPCR function and pharmacology and broadens our understanding of one of the most common pharmaceutical targets to date (8).
Materials and Methods
Methods performed in the current study involved generation of cDNA constructs, cell culture, coimmunoprecipitations, protein G pull-downs, ectodomain purification, amino terminal sequencing by Edman degradation, bioinformatic analysis, and the development of a unique GPCR signaling platform capable of probing functional consequences of transcellular binding partners across intact cells. Using this platform, we utilized numerous luminescence- and bioluminescence resonance energy transfer (BRET)-based biosensors to probe GPCR signaling, including -22F pGloSensor, Nluc-EPAC-VV, and real-time kinetic Gαi/o activation assays, as well as biotinylation membrane expression assays. Extensive details pertaining to these techniques are available in SI Appendix.
Supplementary Material
Acknowledgments
We thank Dr. Ikuo Masuho for guidance with the G protein assays. This work was supported by NIH Grants EY018139 and DA026405 (to K.A.M.). H.A.D. is the recipient of a Canadian Institutes of Health Research Postdoctoral fellowship award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1722498115/-/DCSupplemental.
References
- 1.Sanes JR, Yamagata M. Many paths to synaptic specificity. Annu Rev Cell Dev Biol. 2009;25:161–195. doi: 10.1146/annurev.cellbio.24.110707.175402. [DOI] [PubMed] [Google Scholar]
- 2.Williams ME, de Wit J, Ghosh A. Molecular mechanisms of synaptic specificity in developing neural circuits. Neuron. 2010;68:9–18. doi: 10.1016/j.neuron.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit J, Ghosh A. Specification of synaptic connectivity by cell surface interactions. Nat Rev Neurosci. 2016;17:22–35. doi: 10.1038/nrn.2015.3. [DOI] [PubMed] [Google Scholar]
- 4.Kaeser PS, Regehr WG. Molecular mechanisms for synchronous, asynchronous, and spontaneous neurotransmitter release. Annu Rev Physiol. 2014;76:333–363. doi: 10.1146/annurev-physiol-021113-170338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akil H, et al. Medicine. The future of psychiatric research: Genomes and neural circuits. Science. 2010;327:1580–1581. doi: 10.1126/science.1188654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arguello PA, Gogos JA. Genetic and cognitive windows into circuit mechanisms of psychiatric disease. Trends Neurosci. 2012;35:3–13. doi: 10.1016/j.tins.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Lüthi A, Lüscher C. Pathological circuit function underlying addiction and anxiety disorders. Nat Neurosci. 2014;17:1635–1643. doi: 10.1038/nn.3849. [DOI] [PubMed] [Google Scholar]
- 8.Santos R, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ritter SL, Hall RA. Fine-tuning of GPCR activity by receptor-interacting proteins. Nat Rev Mol Cell Biol. 2009;10:819–830. doi: 10.1038/nrm2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Magalhaes AC, Dunn H, Ferguson SSG. Regulation of GPCR activity, trafficking and localization by GPCR-interacting proteins. Br J Pharmacol. 2012;165:1717–1736. doi: 10.1111/j.1476-5381.2011.01552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn HA, Ferguson SSG. PDZ protein regulation of G protein-coupled receptor trafficking and signaling pathways. Mol Pharmacol. 2015;88:624–639. doi: 10.1124/mol.115.098509. [DOI] [PubMed] [Google Scholar]
- 12.Silva JP, et al. Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci USA. 2011;108:12113–12118. doi: 10.1073/pnas.1019434108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boucard AA, Maxeiner S, Südhof TC. Latrophilins function as heterophilic cell-adhesion molecules by binding to teneurins: Regulation by alternative splicing. J Biol Chem. 2014;289:387–402. doi: 10.1074/jbc.M113.504779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vysokov NV, et al. The mechanism of regulated release of Lasso/teneurin-2. Front Mol Neurosci. 2016;9:59. doi: 10.3389/fnmol.2016.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Sullivan ML, et al. FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron. 2012;73:903–910. doi: 10.1016/j.neuron.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boucard AA, Ko J, Südhof TC. High affinity neurexin binding to cell adhesion G-protein-coupled receptor CIRL1/latrophilin-1 produces an intercellular adhesion complex. J Biol Chem. 2012;287:9399–9413. doi: 10.1074/jbc.M111.318659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niswender CM, Conn PJ. Metabotropic glutamate receptors: Physiology, pharmacology, and disease. Annu Rev Pharmacol Toxicol. 2010;50:295–322. doi: 10.1146/annurev.pharmtox.011008.145533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niciu MJ, Kelmendi B, Sanacora G. Overview of glutamatergic neurotransmission in the nervous system. Pharmacol Biochem Behav. 2012;100:656–664. doi: 10.1016/j.pbb.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masu M, et al. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–765. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- 20.Moldrich RX, Chapman AG, De Sarro G, Meldrum BS. Glutamate metabotropic receptors as targets for drug therapy in epilepsy. Eur J Pharmacol. 2003;476:3–16. doi: 10.1016/s0014-2999(03)02149-6. [DOI] [PubMed] [Google Scholar]
- 21.Lavreysen H, Dautzenberg FM. Therapeutic potential of group III metabotropic glutamate receptors. Curr Med Chem. 2008;15:671–684. doi: 10.2174/092986708783885246. [DOI] [PubMed] [Google Scholar]
- 22.Nicoletti F, et al. Metabotropic glutamate receptors: From the workbench to the bedside. Neuropharmacology. 2011;60:1017–1041. doi: 10.1016/j.neuropharm.2010.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amalric M, et al. Group III and subtype 4 metabotropic glutamate receptor agonists: Discovery and pathophysiological applications in Parkinson’s disease. Neuropharmacology. 2013;66:53–64. doi: 10.1016/j.neuropharm.2012.05.026. [DOI] [PubMed] [Google Scholar]
- 24.Gregory KJ, Noetzel MJ, Niswender CM. Pharmacology of metabotropic glutamate receptor allosteric modulators: Structural basis and therapeutic potential for CNS disorders. Prog Mol Biol Transl Sci. 2013;115:61–121. doi: 10.1016/B978-0-12-394587-7.00002-6. [DOI] [PubMed] [Google Scholar]
- 25.Becker JAJ, et al. Autistic-like syndrome in mu opioid receptor null mice is relieved by facilitated mGluR4 activity. Neuropsychopharmacology. 2014;39:2049–2060. doi: 10.1038/npp.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palazzo E, Marabese I, de Novellis V, Rossi F, Maione S. Metabotropic glutamate receptor 7: From synaptic function to therapeutic implications. Curr Neuropharmacol. 2016;14:504–513. doi: 10.2174/1570159X13666150716165323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolan J, Mitchell KJ. Mutation of Elfn1 in mice causes seizures and hyperactivity. PLoS One. 2013;8:e80491. doi: 10.1371/journal.pone.0080491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tomioka NH, et al. Elfn1 recruits presynaptic mGluR7 in trans and its loss results in seizures. Nat Commun. 2014;5:4501. doi: 10.1038/ncomms5501. [DOI] [PubMed] [Google Scholar]
- 29.de Wit J, Hong W, Luo L, Ghosh A. Role of leucine-rich repeat proteins in the development and function of neural circuits. Annu Rev Cell Dev Biol. 2011;27:697–729. doi: 10.1146/annurev-cellbio-092910-154111. [DOI] [PubMed] [Google Scholar]
- 30.de Wit J, Ghosh A. Control of neural circuit formation by leucine-rich repeat proteins. Trends Neurosci. 2014;37:539–550. doi: 10.1016/j.tins.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sylwestrak EL, Ghosh A. Elfn1 regulates target-specific release probability at CA1-interneuron synapses. Science. 2012;338:536–540. doi: 10.1126/science.1222482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao Y, et al. Mechanism for selective synaptic wiring of rod photoreceptors into the retinal circuitry and its role in vision. Neuron. 2015;87:1248–1260. doi: 10.1016/j.neuron.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, et al. The auxiliary calcium channel subunit α2δ4 is required for axonal elaboration, synaptic transmission, and wiring of rod photoreceptors. Neuron. 2017;93:1359–1374.e6. doi: 10.1016/j.neuron.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- 35.DiRaddo JO, et al. A real-time method for measuring cAMP production modulated by Gαi/o-coupled metabotropic glutamate receptors. J Pharmacol Exp Ther. 2014;349:373–382. doi: 10.1124/jpet.113.211532. [DOI] [PubMed] [Google Scholar]
- 36.Mathiesen JM, Ramirez MT. The metabotropic glutamate receptor 4 is internalized and desensitized upon protein kinase C activation. Br J Pharmacol. 2006;148:279–290. doi: 10.1038/sj.bjp.0706733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aad G, et al. Search for quark contact interactions in dijet angular distributions in pp collisions at root s=7 TeV measured with the ATLAS detector. Phys Lett B. 2011;694:327–345. [Google Scholar]
- 38.Young TR, Leamey CA. Teneurins: Important regulators of neural circuitry. Int J Biochem Cell Biol. 2009;41:990–993. doi: 10.1016/j.biocel.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 39.Gokce O, Südhof TC. Membrane-tethered monomeric neurexin LNS-domain triggers synapse formation. J Neurosci. 2013;33:14617–14628. doi: 10.1523/JNEUROSCI.1232-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.