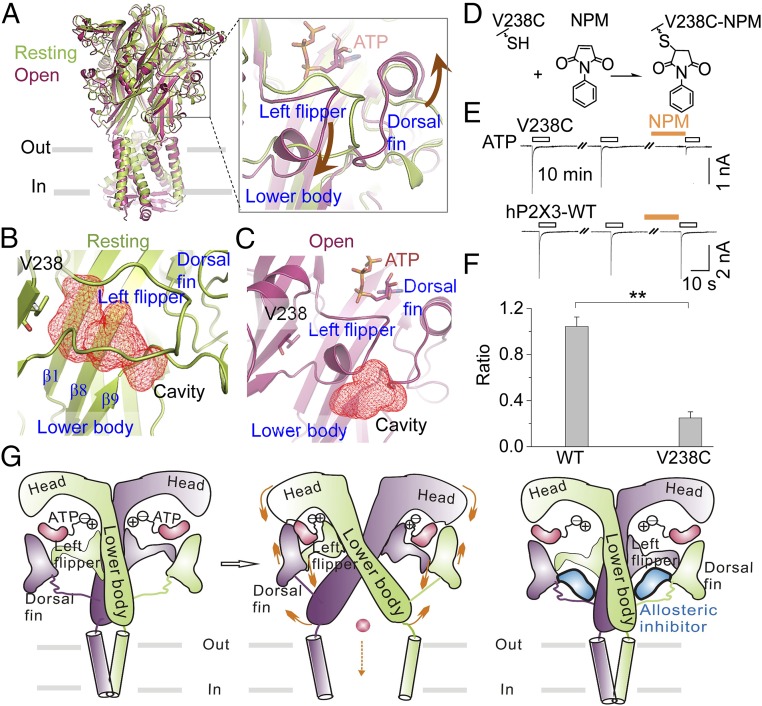
Fig. 5.
Mechanism of allosteric inhibition of P2X3 receptors by the identified site. (A) Superimposition of structures at the resting (lemon, PDB ID code 5SVJ; the missing loop of the LF domain was filled in using homology modeling) and open (warm pink, PDB ID code 5SVK) states of hP2X3 receptors. Brown arrows in the enlarged box indicate the relative motions of the DF and LF domains during ATP-induced channel gating. (B and C) Zoom-in views of cavities, depicted in red mesh for emphasis, fostered by the LF and LB domains of hP2X3 at the resting (B) and open (C) states. (D) Illustration of NPM covalently linked to hP2X3V238C. (E and F) Representative current traces (E) and summary data (F) (n = 4 to 5) for NPM (1 mM) effects on ATP-evoked currents for hP2X3-WT and V238C. **P < 0.01 vs. WT, Student’s t test. (G) Illustrations of the allosteric changes during channel gating of P2X receptors and an allosteric inhibition strategy on P2X3 identified here. Only two chains are shown, for clarity. Orange arrows denote the movements of key domains crucial for channel gating by ATP.