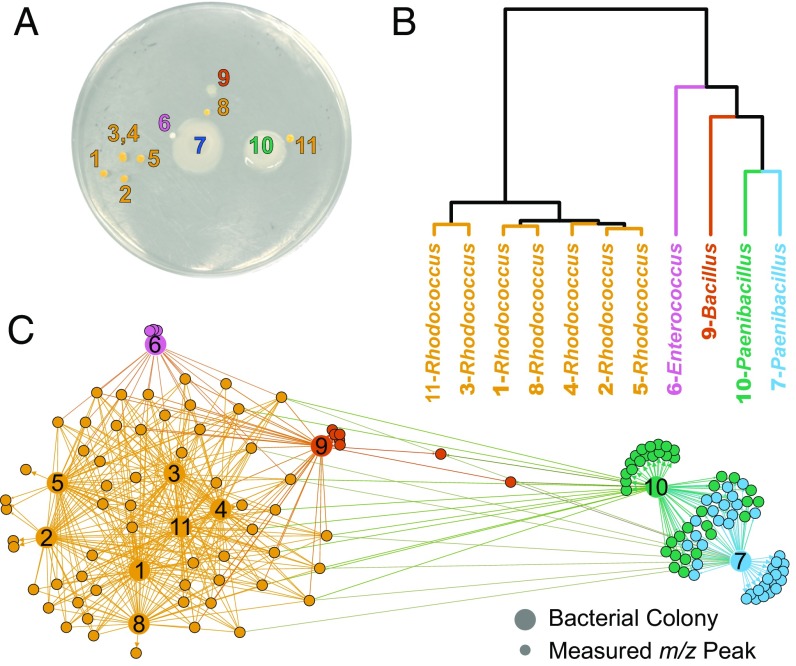
Fig. 3.
Rapid protein and specialized metabolite fingerprinting of unknown environmental isolates using MALDI-TOF MS and IDBac. (A) Bacterial diversity plate obtained from placing freshwater sponge tissue on high-nutrient A1 agar. (B) IDBac allowed for hierarchical clustering of MALDI-TOF MS protein spectra with the option to choose standard distance measures and clustering algorithms. For workflows requiring analysis of hundreds to thousands of strains, protein grouping is essential for data reduction before specialized metabolite analysis is performed. (C) MAN, colored via modularity analysis with default thresholds in Gephi (31), allowed for rapid decision-making based on gross in situ specialized metabolite production after matrix and media signals were subtracted automatically from the network in IDBac. Significant outliers were the two Paenibacillus strains, which produced several shared and unique high molecular-weight specialized metabolites.