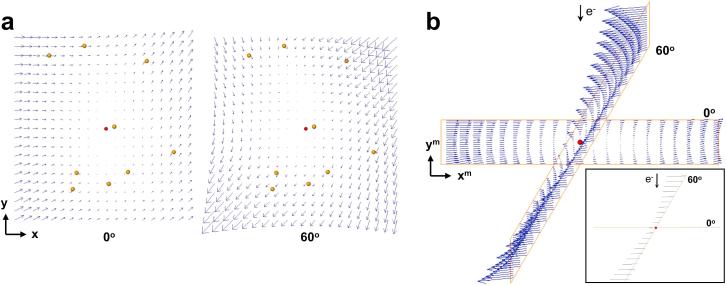
Fig. 2.
Beam-induced motion estimated by the 3D motion-based alignment process. One representative ribosome tilt-series is used as an example. (a) Local 3D shifts given by the polynomial surfaces (bivariate, second degree) that were fitted for the images at 0° (left) and 60° (right). Only the XY components of the vectors are clearly observable in this view (the Z component runs perpendicular to the sheet). Fitted 3D fiducial coordinates are represented with yellow dots. The red dot represents the centroid of the fiducials. The panels show the motion for a field of view of 1 around that centroid. Vectors are magnified by . X- and Y- axes of the sample coordinate system are indicated. (b) Beam-induced motion patterns shown in (a) are presented in the microscope coordinate system, i.e. according to the tilt angle of the views. Note that all the vectors run perpendicular to the electron beam. This is better observed in the inset, which is viewed from the tilt axis and arrowheads have been removed. X- and Y- axes of the microscope coordinate system are indicated and the Z-axis runs along the electron beam direction.