Abstract
Resistance-nodulation-division (RND) superfamily efflux pumps play a major role in multidrug resistance (MDR) of Gram-negative pathogens by extruding diverse classes of antibiotics from the cell. There has been considerable interest in developing efflux pump inhibitors (EPIs) of RND pumps as adjunctive therapies. The primary challenge in EPI discovery has been the highly hydrophobic, poly-specific substrate binding site of the target. Recent findings have identified the hydrophobic trap, a narrow phenylalanine- lined groove in the substrate-binding site, as the “Achilles heel” of the RND efflux pumps. In this review, we will examine the hydrophobic trap as an EPI target and two chemically distinct series of EPIs that bind there.
Keywords: RND superfamily, Efflux pump, Efflux pump inhibitor, Adjunctive therapy, Hydrophobic trap
1. Introduction
The role of the resistance-nodulation-division (RND) superfamily of efflux pumps in the multidrug resistance (MDR) of Gram-negative pathogens is well documented [1]. Efflux pumps in this family contain a polyspecific substrate binding pocket that enables them to bind and extrude a diverse array of compounds from the periplasm to the exterior of the cell, including the majority of clinically used antibiotics. Combined with a highly selective outer membrane, these efflux pumps play a central role in the intrinsic antibiotic resistance of Gram-negative bacteria. RND efflux pumps are present in all Gram-negative pathogens, including Escherichia coli, Klebsiella pneumoniae and Pseudomonas aeruginosa. Pathogenic Gram-negative species encode several distinct RND pumps that contribute significantly to MDR, each with varied substrate specificity and expression patterns [1]. Mutations that upregulate RND efflux pump expression result in increased resistance to multiple classes of antibiotics [2]. In addition, RND efflux pumps play an important role in biofilm formation [3, 4], virulence [5–9] and the emergence of resistance mutations in antibiotic drug targets (e.g. gyrA) [10, 11]. Consequently, there is substantial interest in developing efflux pump inhibitors (EPIs) that target the RND superfamily as adjunctive therapies to be used in combination with antibiotics for infections caused by Gram-negative pathogens [12].
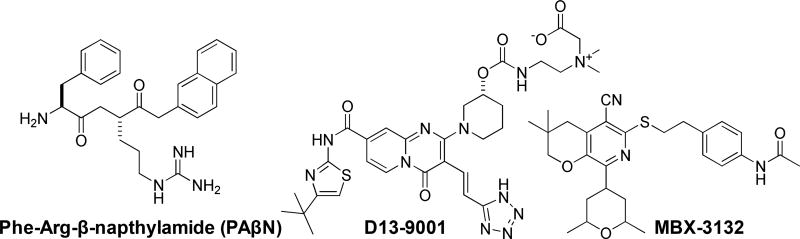
Despite the variety of EPIs reported in the literature [13], few preclinical development programs have been described to date (See Fig. 1). The first notable contribution was from Microcide (later Essential Therapeutics, then Rempex), who developed a family of peptidomimetics, including PAβN (MC-207 110), as an adjunctive therapy with LVX [11, 14–18]. Peptidomimetic EPIs exhibited potent inhibition of efflux pumps in P. aeruginosa and select inhibitors were validated using murine models of infection [15, 16, 18]. However, the program was ultimately put on hold, presumably due to reported nephrotoxicity in the lead series [12]. In 2004, a series of pyridopyrimidine EPIs were reported by a collaborative team from Daiichi Pharmaceuticals and Essential Therapeutics, work that culminated in a preclinical lead compound (D13-9001, Fig. 1)[19–25]. Unfortunately, pyridopyrimidine EPIs exhibited a limited spectrum of activity against P. aeruginosa RND pumps, which limits their clinical applicability and was likely the reason this program was terminated [26]. In 2014, at Microbiotix, we reported the discovery and initial characterization of a pyranopyridine (MBX-2319, Fig. 1) as a potent inhibitor of the major RND efflux pump of E. coli (AcrB) [27]. The pyranopyridine series of EPIs are structurally distinct from the previously reported EPIs, and are active against the pathogens of the Enterobacteriaceae [27, 28], but do not penetrate the outer membrane of P. aeruginosa (unpublished). Optimization of the pyranopyridine series has resulted in the identification of potent compounds that exhibit excellent exposure and tolerability in murine models, and a robust lead optimization program is currently in progress with future preclinical studies planned.
Fig. 1.
Structures of the compounds discussed in this review.
There is a historical perception that EPI drug development is particularly challenging, a viewpoint that is fueled by the lack of major successes in this area;, however, recent developments may be changing that perception. A series of recent reports have identified an “Achilles heel” of the RND efflux pumps, which is likely to spark renewed interest in EPI discovery and development. The primary difficulty in EPI discovery has been that the target is the highly hydrophobic polyspecific substrate binding site of RND efflux pumps. Analyses of previous approaches underscore this hypothesis; the requirement for extensive hydrophobic interactions has resulted in EPIs characterized either by insoluble or amphiphilic molecules, with concomitant issues in pharmacokinetics and toxicity. Additionally, previous efforts have been hampered by issues of selectivity, likely resulting from a limited knowledge of binding pocket interactions. Recently, however, x-ray crystal structures of the pyridopyrimidine [29] and pyranopyridine [30] EPIs bound to the major RND efflux pumps of E. coli and P. aeruginosa have been reported, facilitating a rapid evolution in our understanding of EPI mechanisms and binding interactions. These studies revealed that EPIs bind to a unique site within the substrate binding pocket, known as the hydrophobic trap, with high affinity and do not appear to be pump substrates. The discovery of the hydrophobic trap represents a significant advance in the field, potentially enabling the design of EPIs with increased potency and drug-like properties. In this review, we will examine the recent literature that identified the hydrophobic trap as a target for two chemically distinct series of EPIs, the unique properties of these inhibitors, and potential mechanisms of action. Finally, we will discuss the potential of the hydrophobic trap as a target for EPI drug discovery and the current challenges of EPI drug development.
2.RND superfamily pumps
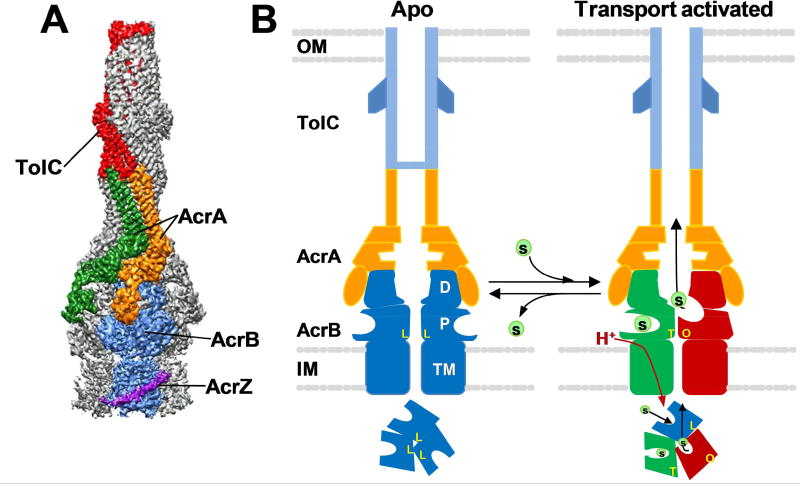
RND superfamily pumps are integral membrane proteins that span the inner membrane (IM), periplasmic space and the outer membrane (OM). AcrABZ-TolC of Escherichia coli is the prototypical RND superfamily pump, as it has been studied most intensively. As implied by its name, AcrABZ-TolC is comprised of four proteins, each with a distinct function (see Fig. 2A). AcrB is an integral membrane protein that protrudes into the periplasmic space and is a permease that uses proton motive force to drive the process of efflux. TolC is a porin-like protein that extends from the periplasmic space and traverses the outer membrane. AcrA is a so-called membrane fusion protein (MFP) because it connects AcrB and TolC and transmits conformational changes from AcrB to TolC [18–20]. AcrZ is a highly conserved 49 amino acid protein [31] that interacts with the transmembrane domain of AcrB [32] and affects substrate recognition [31]. Recent cryo-EM structures of the entire AcrABZ-TolC complex confirmed the overall architecture of the pump and the subunit stoichiometry of 3 AcrB: 6 AcrA: 3 AcrZ: 3 TolC [33] (see Fig. 2A). Other orthologous RND pumps that extrude antibiotics, such as MexAB-OprM of P. aeruginosa, share the same architecture as AcrAB-TolC.
Fig. 2.
Structure and transport mechanism of the AcrABZ-TolC RND efflux pump. A) A cryo-EM structure of AcrABZ-TolC bound to MBX-3132 with a resolution of 3.6 Å. The entire complex is comprised of 3 AcrB, 3 AcrZ, 6 AcrA and 3 TolC subunits. The components of a single functional unit of the trimeric pump are labeled. This image was reproduced with permission from Wang et al. [33]. B) A cartoon depicting the current model of the RND efflux pump transport mechanism. In the absence of substrate (right panel), the pump is in a resting (apo) state, in which the AcrB trimer is in the LLL conformation and the central channel of TolC is closed. In the presence of efflux substrates (left panel), the pump switches to a transport activated state characterized by changes in AcrB that result in three distinct conformation states (L, T and O), which represent discreet stages of the transport mechanism. Each subunit cycles from the L to T to O conformation, a process that is driven by the translocation of protons through the transmembrane domain (TM) of AcrB and is essential for unidirectional transport. When the substrates have been depleted, the pump reverts to the apo state. The cartoons depict cross-section views through the length of the pump in which two protomers of each of the pump components are shown. The cartoons below depict a view of the molecular axis of the AcrB trimer, which shows conformations of the AcrB protomers in the apo and transport activated states. Key: OM, outer membrane; IM, inner membrane; TM, AcrB transmembrane domain; P, AcrB pore domain; D, AcrD docking domain; L, T and O, Loose, Tight and Open conformations, respectively, of AcrB. This cartoon was adapted from a figure published by Wang et al. [33].
3.Structure and function of AcrB
The current model for the structure and function of the AcrABZ-TolC efflux pump is shown in Fig. 2B. AcrB is comprised of three distinct domains (reviewed in [2, 34–36]): 1) a transmembrane (TM) domain comprised of 12 transmembrane helices, which uses proton motive force to drive the efflux process [37–39]; 2) a porter (P) domain comprised of the two large periplasmic loops that bind and extrude substrates; and 3) a dock (D) domain that binds to AcrA. AcrB assumes three distinct conformations during the efflux process, known as Loose (L), Tight (T) and Open (O) [40–42], or Access, Binding and Extrusion [40], respectively. In this review, we will refer to the conformations as L, T and O. In the absence of substrates, the AcrB homotrimer exists in an Apo form, in which all protomers are in the L conformation [33]. When substrates are present, AcrB undergoes conformational changes to form an asymmetric trimer that is transport activated [33]. The activated asymmetric trimer comprises one protomer each in the L, T and O conformations [33, 35, 40, 43]. Several lines of evidence indicate that each conformation represents a discreet stage of the efflux process [39, 44, 45]. In the current model, each subunit successively assumes each of the conformations as substrates first interact with the L conformation at the periplasmic access binding site. The substrate is moved to the deep substrate binding pocket in the T conformation. Transport of protons through the TM domain drives the transition to the O conformation, which is characterized by the collapse of the deep substrate binding pocket, causing the substrate to be extruded into a central channel that leads to TolC. The T to O transition and release of the substrate only occurs when a substrate is bound to the adjacent L protomer [43]. This model suggests that the conformational changes within the LTO trimer are coordinated by intersubunit interactions, which predicts that one defective protomer will inactivate the entire trimeric pump. This prediction was verified by an experiment demonstrating that a single inactive protomer defective in proton translocation in a covalently-linked AcrB trimer inactivated the entire pump [39].
4.The substrate binding site of AcrB
The substrate binding pocket of AcrB is polyspecific and is able to bind and pump a diverse array of chemicals, including many clinically important antibiotics, which complicates the discovery and design of potent, specific EPIs. Therefore, the structure of the substrate binding pocket in the T protomer, also known as the distal binding site, is particularly relevant to EPI discovery and design. The substrate binding pocket is clearly defined in the three-dimensional crystal structure of an AcrB protomer in the T conformation bound to the pump substrates minocycline (MIN) and doxorubicin (DOX) [40]. These structures revealed that the binding pocket is a large cavity lined with hydrophobic (F136 and F178, F610, F615, F617 and F628) and polar residues (N274 and Q176). Both compounds interacted mainly with hydrophobic residues through Van der Waals and ring stacking interactions and made hydrogen bonding interactions with the polar residues. Interestingly, MIN and DOX interacted with a distinct set of amino acid residues, providing an explanation for the polyspecificity of the pump, which was supported by the results of docking and molecular dynamic simulation studies [46, 47]. Substrate binding is characterized by low binding affinity and interactions based on generalized physical properties, such as hydrophobicity, resulting in a broad diversity of bound substrates. Consequently, the substrate binding site is a challenging target for EPI discovery and development, as competitive inhibitors are likely to have properties similar to those of substrates. This is exemplified by the EPI PAβN (Fig. 1), which is likely to bind within the substrate binding pocket. PAβN has been shown to be a substrate of the RND pumps [11, 48] that binds to AcrB with low affinity, as demonstrated by the results of a quantitative cell-based efflux assay (Km 17.6 ± 5.0 µM) [48] and by a surface plasmon resonance binding assay (Kd 15 – 28 µM) [49], and was supported by molecular dynamic simulations [47]. The antibiotic potentiating activity of this EPI is strongly dependent on the properties (binding site) of the antibiotic [11], suggesting that PAβN inhibits efflux by competing with substrates for binding and transport from overlapping sites in the binding pocket.
5.The hydrophobic trap
The hydrophobic trap is a unique binding site within the substrate binding pocket of RND pumps. This site is a narrow cleft lined with hydrophobic residues (F136, F178, F610, F615 and F628), which inspired its name [29]. The hydrophobic trap was identified when the first 3D crystal structures of an efflux pump inhibitor (D13-9001) bound to AcrB and MexB were reported by Nakashima et al. in 2013 [29]. The hydrophobic tert-butyl thiazolyl aminocarboxyl pyridopyrimidine (TAP) moiety of D13-9001 (see Fig. 1) interacts with F178 and F628 through π-π interactions. The tetrazole ring and the piperidine aceto-amino ethylene ammonioacetate (PAEA) moieties of D13-9001 interact with ionic and/or hydrophilic residues (N274, R620, Q176 and S180) and with aliphatic residues (I277 and L177) that are in the substrate translocation channel, and extends into the substrate binding sites of both MIN and DOX. The binding site of D13-9001 in MexB was similar to the binding site in AcrB, except for differences in the binding of the PAEA moiety in the substrate translocation channel. D13-9001 binds to AcrB and MexB with KD values of 1.15 µM and 3.57 µM, respectively, and does not appear to be exported by either pump [29].
The importance of the hydrophobic trap was confirmed when Sjuts et al. reported the 3D structures of a series of potent pyranopyridine EPIs bound to AcrBper [30], an engineered protein comprised of the periplasmic domains (porter and docking) of AcrB that is structurally identical to the full-length protein. In the co-crystal structure, the pyranopyridines bind to the hydrophobic trap of the T protomer, where they engage in multiple hydrophobic interactions with side chains lining the deep binding pocket and hydrophobic trap. The central, aromatic pyridine ring is oriented parallel to the F628 aromatic side chain, resulting in an extensive π–π stacking interaction. Similarly, the phenyl and morpholinyl substituents of the central ring interact with F178 and F615. The F610 side chain is orthogonally packed against the dimethylene sulfide moiety that connects the pyranopyridine core to the phenyl group. The side chains of Y327 and M573 interact with the gem dimethyl group. In addition, the acetamide of MBX-3132 was found to exhibit interactions with AcrB through a complicated network of hydrogen bonds centered on a solvent water that is highly coordinated by the conserved A286 carbonyl backbone oxygen and the Q151 side chain. The dimethylmorpholinyl group protrudes into the substrate binding pocket and forms a hydrogen bond with a water molecule coordinate by Q176. Like D13-9001, MBX-3132 is a potent inhibitor of AcrB that completely inhibits efflux at a concentration of 10 nM [30]. Currently, it is not known whether MBX-3132 is a substrate of AcrB.
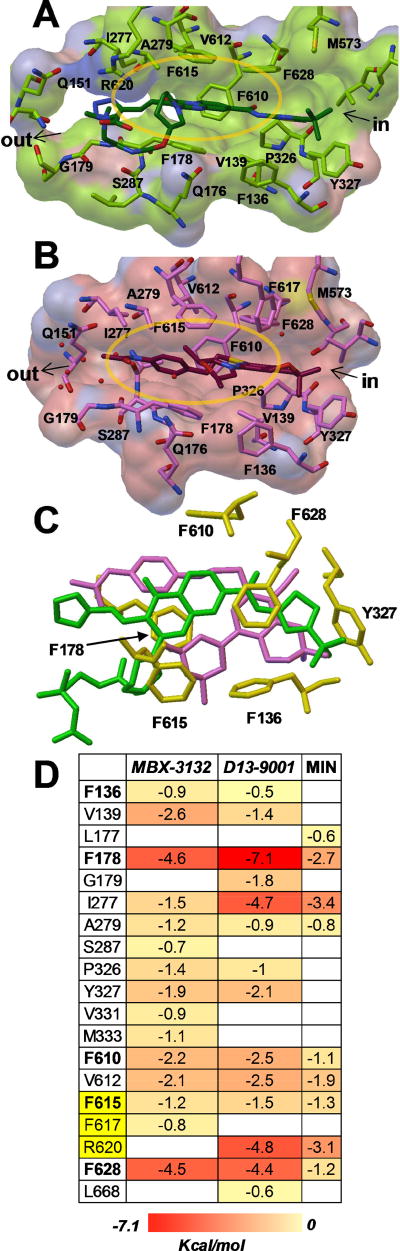
Comparison of the binding sites of MBX-3132 and D13-9001 reveal additional features. The bound structures of the two compounds exhibit significant overlap and interact with a highly correlated set of residues (Fig. 3C), as illustrated by MD simulations that quantified interactions between the EPIs and amino acid residues in and around the hydrophobic trap (Fig. 3D) [30]. Both compounds make strong hydrophobic interactions with F178 and F628 and extend polar groups into the water filled pocket near the exit of the trap (near Q151). Additionally, there is a close overlap between the tert-butyl group of D13-9001 and the gem-dimethyl group of MBX-3132 near Y327. Finally, both compounds make hydrogen bonds to Q176 (MBX-3132 through a water bridge with the morpholine oxygen, D13-9001 through the carbamate oxygen). The interactions of the polar substituents near Q151 and Q176 and the hydrophobic groups at Y327 may anchor the compounds in position, potentially driving the specificity and potency seen in both series.
Fig. 3.
Potent efflux pump inhibitors D13-9001 and MBX-3132 bound to the hydrophobic trap of AcrB. A) D13-9001 (green, carbon; red, oxygen; blue, nitrogen; PDB entry 3W9H [29]). B) MBX-3132 (dark red, carbon; PDB entry 5ENQ [30]). The surfaces have been colored green and pink to highlight residues within 5 Å of D13-9001 and MBX-3132, respectively. Images were generated with Pymol [51]. C) Superimposition of MBX-3132 (purple, PDB entry 5ENO [30]) and D13-9001 (green, PDB entry 5ENO [30]) bound to the hydrophobic trap of AcrB shows the differences and similarities of their binding sites. Amino acid side chains of residues comprising the hydrophobic trap are shown as yellow sticks. Image was generated with Pymol [51]. D) The relative per-residue contributions to the free energy of binding (ΔGb, kilocalories per mole) were calculated for D13-9001 and MBX-3132 bound to AcrB, and are compared with estimates calculated for MIN [43]. Residues comprising the hydrophobic trap [29] are shown in bold. Residues in the switch loop are highlighted in yellow. The data for MBX-3132 were reprinted from Sjuts et al. [30] with permission and the data for D13-9001 were kindly provided by Attilio Vargiu (University of Cagliari).
In addition to the similarities between the binding interactions of D13-9001 and MBX-3132 with AcrB, there are a few notable differences. The bicyclic core of D13-9001 appears to maintain stronger π-π interactions with F178 than the pendant phenyl moiety of MBX-3132. Similarly, the glycine sidechain of D13-9001 maintains a salt bridge with R620 that is entirely lacking in MBX-3132. In contrast, the through-water hydrogen bonding network between the acetamide of MBX-3132, Q151, S287 and S155 appears to be more direct than the polar interactions of the tetrazole in D13-9001. The distinctions in binding interactions provides potential insights and strategies for future iterations of both of these promising series, as well as potentially providing key strategies for future EPI scaffolds.
6.Conservation of the hydrophobic trap
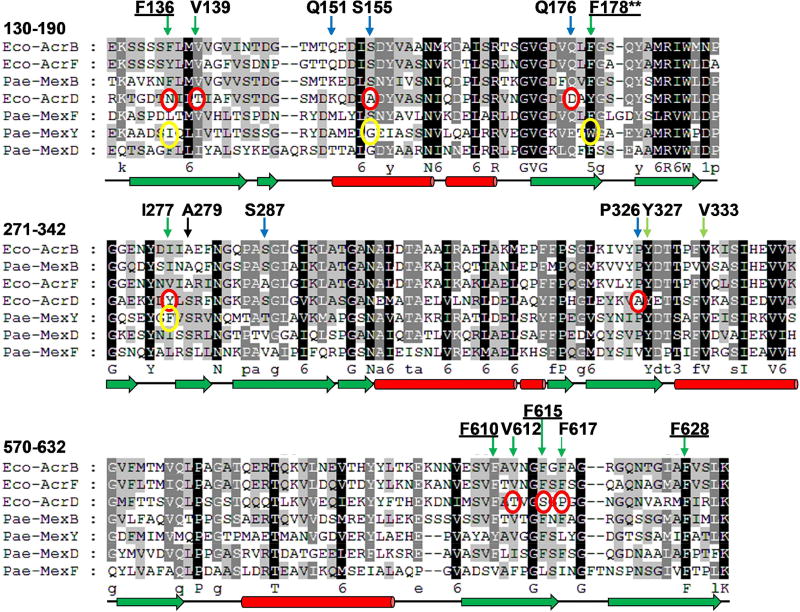
The amino acid residues that comprise the hydrophobic trap of AcrB orthologs in the Enterobacteriaceae are highly conserved (data not shown). However, as shown in Fig. 4, a comparison of the hydrophobic trap from the RND pumps in E. coli (AcrB, AcrD, AcrF) and P. aeruginosa (MexB, MexD, MexF and MexY) that transport antibiotics reveals significant differences in the hydrophobic trap residues of AcrD and MexY, as compared to the other pumps, with the exception of F628 (which is highly conserved). Substrates of AcrD and MexY differ significantly from the other pumps, and include hydrophilic antibiotics, such as aminoglycosides. As mentioned above, D13-9001 is active against MexB/AcrB, but not MexY [29], which has a Trp (W) residue at position 178 instead of Phe (F), as is present in the other pumps. A homology model of the three dimensional structure of MexY predicted that D13-9001 would not bind to the hydrophobic trap because the F178W substitution would prevent binding by steric hindrance. This model was confirmed when a site-directed mutant of MexY (W177F) was shown to be sensitive to D13-9001, and the converse substitution in AcrB (F178W) does not bind the EPI [29]. It is not known whether D13-9001 is active against AcrD; however, AcrD does not have the F178W substitution, so it is possible that this compound will inhibit the aminoglycoside pump of E. coli. Ramaswamy et al. constructed a homology model of the three-dimensional structure of AcrD and carried out extensive molecular dynamic simulations comparing the substrate binding pockets of AcrB and AcrD, including the hydrophobic trap [50]. Their results indicate that the numerous substitutions of hydrophobic residues by hydrophilic residues in AcrD drastically alter the physicochemical properties of the hydrophobic trap. In addition, substitutions in the switch loop of AcrD are predicted to alter substrate recognition and transport, and possibly interactions with inhibitors (see below). Preliminary evidence from our lab indicates that MBX-3132 is active against AcrB and AcrF, but not AcrD (manuscript in preparation). Interestingly, several residues in AcrB that interact with MBX-3132 have non-conservative substitutions in AcrD, including F136N and F615S. Experiments in our lab are under way to determine the effects of these substitutions on the EPI activity of MBX-3132. Therefore, the divergence of the hydrophobic trap of AcrD/MexY from AcrB/MexB is a major challenge for the design of EPIs that are active against all of the major RND superfamily pumps, especially as structural characterization of AcrD or MexY has not been reported. While a homology model of AcrD [50] would be useful for evaluating novel inhibitors, a three-dimensional structure of one of both of these pumps would enable the use of structure-based drug design methods to improve the spectrum of activity of existing EPIs.
Fig. 4.
A multi-sequence alignment of the major RND superfamily pumps of E. coli and P. aeruginosa showing conservation of the residues that comprise the hydrophobic trap. Residues that interact with MBX-3132 and D13-9001 are indicated above the alignment with green and blue arrows to indicate hydrophobic and polar residues, respectively. Hydrophobic trap residues are underlined. The residues are numbered according to the E. coli AcrB sequence. Significant differences between the conserved residues of AcrB/MexB and AcrD and MexY are indicated by red and yellow circles, respectively. Residue F178 is highlighted by two asterisks (**) because the F178W substitution in MexY has been shown to prevent binding of D13-9001 [29]. The secondary structure of AcrB is shown below the alignment using green arrows to indicate beta strands and red cylinders to indicate alpha helices. The alignment was generated using Clustal X2 [52].
7.Mechanistic implications of the hydrophobic trap
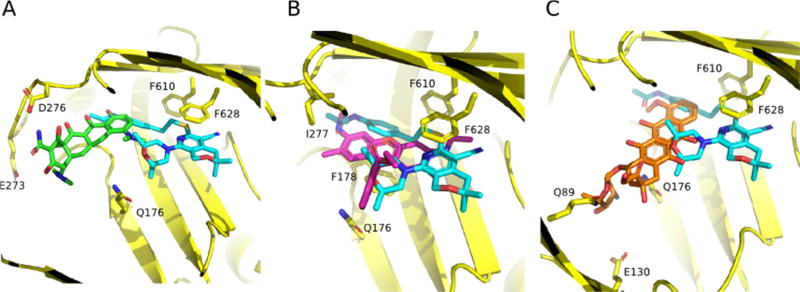
The mechanism of inhibition of EPIs that bind to the hydrophobic trap is still unclear, although there are several possibilities that are not mutually exclusive. The first and most obvious mechanism is one of steric hindrance. As shown in Fig. 5, the binding site of MBX-3132 extends into the substrate binding pocket of AcrB and overlaps with the binding sites of known substrates minocycline, R6G and doxorubicin [30]. Similarly, the binding site of D13-9001 extends into the substrate binding pocket and also overlaps with the binding sites for minocycline and doxorubicin [29]. It is likely that these EPIs bind to the hydrophobic trap with higher affinity than efflux substrates and are able to block substrate binding.
Fig. 5.
The MBX compound binding site overlaps with substrate binding sites. Superimposition of MBX-3132 (cyan, carbon; red, oxygen; blue, nitrogen; yellow, sulfur) bound to AcrB with substrate compounds. A) MIN (green; PDB entry 4DX5 [43]), B) R6G (magenta; PDB entry 5ENS [30]) and C) doxorubicin (orange; PDB entry 4DX7 [43]). MBX-3132 bound to the hydrophobic trap sterically hinders substrate binding to the AcrB deep binding pocket. AcrB side-chains involved in the binding of substrates or EPI are indicated and shown as sticks (carbon = yellow). Reprinted with permission from Sjuts et al. [30].
High affinity binding to the hydrophobic trap suggests a second mechanism in which the EPI freezes the AcrB trimer in an inactive conformation. When cryo-EM was used to determine the structure of MBX-3132 bound to AcrABZ-TolC, 73% of the complexes comprised AcrB trimers that were in the TTT conformation [33]. The authors of the paper suggested that this finding indicates that MBX-3132 binds to AcrB with high affinity [33]. A similar observation was made by the Pos laboratory: when AcrB is crystallized in the presence of high concentrations of substrate, AcrB trimers are in the TTT conformation [34]. These findings indicate that the substrate binding sites in the AcrB trimer can be saturated, which disrupts the conformation of the functionally rotating AcrB trimer (LTO conformation). The fate of the TTT trimers is not known. It is possible that lower affinity substrates could be transported and the AcrB trimer could resume efflux according to the current functional rotation model. However, this would require each subunit to act independently, which has not been observed experimentally. It is likely that high-affinity EPIs that bind to the hydrophobic trap are not readily removed by efflux, effectively locking the AcrB trimer in the TTT conformation and disrupting the inter-subunit interactions that coordinate conformational changes during normal pump function.
A third possible mechanism is that high affinity binding to the hydrophobic trap prevents conformational changes required for the proper activity of the pump. Nakashima et al. concluded that D13-9001 binds tightly to the hydrophobic trap and prevents the conformational changes that are needed for the proper activity of the pump [29]. Indeed, it is possible that MBX-3132 and D13-9001 prevent the movement of the so-called “switch loop,” a flexible loop that separates the access and the deep substrate binding sites in the L and T conformations, respectively [43]. Conformational changes of the switch loop are required for the movement of substrates from the access site to the deep substrate binding pocket [43]. Based on the results of molecular dynamic simulations of the EPIs PAβN and NMP binding the substrate binding pocket of AcrB, Vargiu and Nikaido first proposed that these EPIs inhibit the conformational changes required for the movement of substrates through the pump by interacting with residues in the switch loop [47]. The co-crystal structure of MBX-3132 bound to AcrB, and the results of molecular dynamic simulations demonstrate that MBX-3132 interacts with F615 and F617 of the switch loop [30] (Fig. 3D). In addition, the co-crystal structure of D13-9001 bound to AcrB shows a strong interaction with R620 of the switch loop [29] (Fig. 3D). Therefore, it is possible that both of these EPIs inhibit the movement of substrates into the deep substrate binding pocket.
8.Challenges
The development of a successful EPI inhibitor faces two key challenges beyond those common to antibiotic development: it is obligated to be an adjunctive therapy, and the compound binding site is highly hydrophobic. The development of an adjunctive therapy requires careful titration of pharmacokinetic parameters, including both half-life and distribution to ensure efficacy as well as consideration of drug-drug interactions (Cyp inhibition and synergistic side effects among others). Additional considerations will occur when considering general applications – the ideal candidate must work with a range of antibiotics to maximize effect against varying pathogens and strains, adding further pharmacokinetic constraints into the drug development process. The extremely hydrophobic nature of the AcrB binding site has historically limited effective EPIs to either insoluble or amphiphilic molecules, with concomitant issues in pharmacokinetics and toxicity. As improvements in understanding molecular interactions at this site progress, new strategies will need to emerge that improve physical properties of EPIs and, ultimately, abrogate this barrier to development.
Acknowledgments
We would like to thank Attilio Vargiu (University of Cagliari) for kindly providing the unpublished data shown in Fig. 3D. This work was supported by the National Institute of Allergy and Infectious Diseases (Grant R44 AI100332).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li XZ, Plesiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28:337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nikaido H, Pages JM. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol Rev. 2012;36:340–63. doi: 10.1111/j.1574-6976.2011.00290.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kvist M, Hancock V, Klemm P. Inactivation of efflux pumps abolishes bacterial biofilm formation. Appl Environ Microbiol. 2008;74:7376–82. doi: 10.1128/AEM.01310-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y, Yang L, Molin S. Synergistic activities of an efflux pump inhibitor and iron chelators against Pseudomonas aeruginosa growth and biofilm formation. Antimicrob Agents Chemother. 2010;54:3960–3. doi: 10.1128/AAC.00463-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blair JM, La Ragione RM, Woodward MJ, Piddock LJ. Periplasmic adaptor protein AcrA has a distinct role in the antibiotic resistance and virulence of Salmonella enterica serovar Typhimurium. J Antimicrob Chemother. 2009;64:965–72. doi: 10.1093/jac/dkp311. [DOI] [PubMed] [Google Scholar]
- 6.Hirakata Y, Kondo A, Hoshino K, Yano H, Arai K, Hirotani A, et al. Efflux pump inhibitors reduce the invasiveness of Pseudomonas aeruginosa. Int J Antimicrob Agents. 2009;34:343–6. doi: 10.1016/j.ijantimicag.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 7.Nishino K, Latifi T, Groisman EA. Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol Microbiol. 2006;59:126–41. doi: 10.1111/j.1365-2958.2005.04940.x. [DOI] [PubMed] [Google Scholar]
- 8.Ricci V, Piddock LJ. Exploiting the role of TolC in pathogenicity: identification of a bacteriophage for eradication of Salmonella serovars from poultry. Appl Environ Microbiol. 2010;76:1704–6. doi: 10.1128/AEM.02681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Virlogeux-Payant I, Baucheron S, Pelet J, Trotereau J, Bottreau E, Velge P, et al. TolC, but not AcrB, is involved in the invasiveness of multidrug-resistant Salmonella enterica serovar Typhimurium by increasing type III secretion system-1 expression. Int J Med Microbiol. 2008;298:561–9. doi: 10.1016/j.ijmm.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Lomovskaya O, Lee A, Hoshino K, Ishida H, Mistry A, Warren MS, et al. Use of a genetic approach to evaluate the consequences of inhibition of efflux pumps in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1999;43:1340–6. doi: 10.1128/aac.43.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lomovskaya O, Warren MS, Lee A, Galazzo J, Fronko R, Lee M, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45:105–16. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic--a vision for applied use. Biochem Pharmacol. 2006;71:910–8. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Van Bambeke F, Pages JM, Lee VJ. Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat Antiinfect Drug Discov. 2006;1:157–75. doi: 10.2174/157489106777452692. [DOI] [PubMed] [Google Scholar]
- 14.Renau TE, Leger R, Filonova L, Flamme EM, Wang M, Yen R, et al. Conformationally-restricted analogues of efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2003;13:2755–8. doi: 10.1016/s0960-894x(03)00556-0. [DOI] [PubMed] [Google Scholar]
- 15.Renau TE, Leger R, Flamme EM, Sangalang J, She MW, Yen R, et al. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J Med Chem. 1999;42:4928–31. doi: 10.1021/jm9904598. [DOI] [PubMed] [Google Scholar]
- 16.Renau TE, Leger R, Flamme EM, She MW, Gannon CL, Mathias KM, et al. Addressing the stability of C-capped dipeptide efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2001;11:663–7. doi: 10.1016/s0960-894x(01)00033-6. [DOI] [PubMed] [Google Scholar]
- 17.Renau TE, Leger R, Yen R, She MW, Flamme EM, Sangalang J, et al. Peptidomimetics of efflux pump inhibitors potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorg Med Chem Lett. 2002;12:763–6. doi: 10.1016/s0960-894x(02)00006-9. [DOI] [PubMed] [Google Scholar]
- 18.Watkins WJ, Landaverry Y, Leger R, Litman R, Renau TE, Williams N, et al. The relationship between physicochemical properties, in vitro activity and pharmacokinetic profiles of analogues of diamine-containing efflux pump inhibitors. Bioorg Med Chem Lett. 2003;13:4241–4. doi: 10.1016/j.bmcl.2003.07.030. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama K, Ishida Y, Ohtsuka M, Kawato H, Yoshida K, Yokomizo Y, et al. MexAB-OprM-specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 1: discovery and early strategies for lead optimization. Bioorg Med Chem Lett. 2003;13:4201–4. doi: 10.1016/j.bmcl.2003.07.024. [DOI] [PubMed] [Google Scholar]
- 20.Nakayama K, Ishida Y, Ohtsuka M, Kawato H, Yoshida K, Yokomizo Y, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 2: achieving activity in vivo through the use of alternative scaffolds. Bioorg Med Chem Lett. 2003;13:4205–8. doi: 10.1016/j.bmcl.2003.07.027. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama K, Kawato H, Watanabe J, Ohtsuka M, Yoshida K, Yokomizo Y, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 3: Optimization of potency in the pyridopyrimidine series through the application of a pharmacophore model. Bioorg Med Chem Lett. 2004;14:475–9. doi: 10.1016/j.bmcl.2003.10.060. [DOI] [PubMed] [Google Scholar]
- 22.Nakayama K, Kuru N, Ohtsuka M, Yokomizo Y, Sakamoto A, Kawato H, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 4: Addressing the problem of poor stability due to photoisomerization of an acrylic acid moiety. Bioorg Med Chem Lett. 2004;14:2493–7. doi: 10.1016/j.bmcl.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida K, Nakayama K, Kuru N, Kobayashi S, Ohtsuka M, Takemura M, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 5: Carbon-substituted analogues at the C-2 position. Bioorg Med Chem. 2006;14:1993–2004. doi: 10.1016/j.bmc.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida K, Nakayama K, Ohtsuka M, Kuru N, Yokomizo Y, Sakamoto A, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem. 2007;15:7087–97. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 25.Yoshida K, Nakayama K, Yokomizo Y, Ohtsuka M, Takemura M, Hoshino K, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 6: exploration of aromatic substituents. Bioorg Med Chem. 2006;14:8506–18. doi: 10.1016/j.bmc.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 26.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Opperman TJ, Kwasny SM, Kim HS, Nguyen ST, Houseweart C, D'Souza S, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014;58:722–33. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen ST, Kwasny SM, Ding X, Cardinale SC, McCarthy CT, Kim HS, et al. Structure-activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg Med Chem. 2015;23:2024–34. doi: 10.1016/j.bmc.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakashima R, Sakurai K, Yamasaki S, Hayashi K, Nagata C, Hoshino K, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500:102–6. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 30.Sjuts H, Vargiu AV, Kwasny SM, Nguyen ST, Kim HS, Ding X, et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc Natl Acad Sci U S A. 2016;113:3509–14. doi: 10.1073/pnas.1602472113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobbs EC, Yin X, Paul BJ, Astarita JL, Storz G. Conserved small protein associates with the multidrug efflux pump AcrB and differentially affects antibiotic resistance. Proc Natl Acad Sci U S A. 2012;109:16696–701. doi: 10.1073/pnas.1210093109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du D, Wang Z, James NR, Voss JE, Klimont E, Ohene-Agyei T, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509:512–5. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Fan G, Hryc CF, Blaza JN, Serysheva II, Schmid MF, et al. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. eLife. 2017;6 doi: 10.7554/eLife.24905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794:782–93. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Eicher T, Brandstatter L, Pos KM. Structural and functional aspects of the multidrug efflux pump AcrB. Biol Chem. 2009;390:693–9. doi: 10.1515/BC.2009.090. [DOI] [PubMed] [Google Scholar]
- 36.Blair JM, Piddock LJ. Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr Opin Microbiol. 2009;12:512–9. doi: 10.1016/j.mib.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Seeger MA, von Ballmoos C, Verrey F, Pos KM. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry. 2009;48:5801–12. doi: 10.1021/bi900446j. [DOI] [PubMed] [Google Scholar]
- 38.Su CC, Li M, Gu R, Takatsuka Y, McDermott G, Nikaido H, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188:7290–6. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takatsuka Y, Nikaido H. Covalently linked trimer of the AcrB multidrug efflux pump provides support for the functional rotating mechanism. J Bacteriol. 2009;191:1729–37. doi: 10.1128/JB.01441-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–9. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 41.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–8. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 42.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5:e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eicher T, Cha HJ, Seeger MA, Brandstatter L, El-Delik J, Bohnert JA, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci U S A. 2012;109:5687–92. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eicher T, Seeger MA, Anselmi C, Zhou W, Brandstatter L, Verrey F, et al. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 2014;3 doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seeger MA, von Ballmoos C, Eicher T, Brandstatter L, Verrey F, Diederichs K, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15:199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 46.Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci U S A. 2010;107:6559–65. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci U S A. 2012;109:20637–42. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kinana AD, Vargiu AV, May T, Nikaido H. Aminoacyl beta-naphthylamides as substrates and modulators of AcrB multidrug efflux pump. Proc Natl Acad Sci U S A. 2016;113:1405–10. doi: 10.1073/pnas.1525143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mowla R, Wang Y, Ma S, Venter H. Kinetic analysis of the inhibition of the drug efflux protein AcrB using surface plasmon resonance. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbamem.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 50.Ramaswamy VK, Vargiu AV, Malloci G, Dreier J, Ruggerone P. Molecular Rationale behind the Differential Substrate Specificity of Bacterial RND Multi-Drug Transporters. Scientific reports. 2017;7:8075. doi: 10.1038/s41598-017-08747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanner MF, Duncan BS, Carrillo CJ, Olson AJ. Integrating computation and visualization for biomolecular analysis: an example using python and AVS. Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing. 1999;401:12. doi: 10.1142/9789814447300_0039. [DOI] [PubMed] [Google Scholar]
- 52.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]