Abstract
The potential for local radiation therapy to elicit systemic (abscopal) anti-tumor immune responses has been receiving a significant amount of attention over the last decade. We recently developed a mathematical framework designed to simulate the systemic dissemination of activated T cells among multiple metastatic sites. This framework allowed the identification of non-intuitive patterns of T cell redistribution after localized therapy, and offered suggestions as to the optimal site to irradiate in order to increase the magnitude of an immune-mediated abscopal response. Here, we evaluate the potential for such a framework to provide clinical decision making support to radiation oncologists. Several challenges such as efficient segmentation and delineation of multiple tumor sites on PET/CT scans, validation of model prediction performance, and effective clinical trial design remain to be addressed prior to the incorporation of such a tool in the clinical setting.
1. Introduction
It is well established that antigen presenting cells (APCs) can activate naive T cells in the tumor-draining lymph nodes after encountering tumor-associated antigens [1]. Newly activated T cells subsequently proliferate and migrate through the blood system to the tumor site to exert their cytotoxic effects [2]. Localized radiation therapy can induce immunogenic cell death, exposing pre-existing and de novo tumor-associated antigens and danger-associated molecular patterns [3, 4]. This allows the activation of a greater number of APCs, which can then prime more T cells to be disseminated and attack the tumor. Thus, radiation is being increasingly accepted as a valuable tool for boosting the antitumor immune response.
In the presence of several metastatic sites, the dissemination patterns of T cells after local activation in the tumor draining lymph node are less intuitive. Recently, Poleszczuk and colleagues developed a mathematical framework for predicting this activated T cell dissemination based on T cell trafficking through the circulatory system [5]. Several factors, including blood flow fraction to each tumor and metastasis-bearing organ and T cell tendency to extravasate from the blood at the site of initial APC activation, were assumed to influence T cell redistribution. The model allows the quantification of a newly defined Immunogenicity Index; each respective metastatic site can be assigned a score representing the systemic impact of T cells activated therein.
Simulations of virtual patients with a wide range of potential metastatic tumor distributions suggested that not all metastatic sites participate in systemic immune surveillance equally, and confirm that activated T cell redistribution between sites is highly non-intuitive. The authors conclude that certain metastatic sites with higher immunogenicity indices may have greater potential to induce a systemic (abscopal) anti-tumor immune response in distant metasases, and thus may be optimal sites for targeted local irradiation [5]. The sites inducing the most beneficial redistribution of activated T cells among metastases did not necessarily correspond to the obvious clinical choice of target site, for example the tumor with largest volume.
The development of such simple and tractable quantitative models based on fundamental principles of cancer biology and immunology can isolate and investigate key mechanisms in complex biological processes and evaluate their role in the broader setting of patient response [6, 7, 8]. However, the ability of such a framework to aid decision making in the clinical setting relies on accurate and efficient data collection and analysis, appropriately calibrated models, computationally efficient simulations, thorough prediction performance analysis, and ultimately validation through clinical trials. Here, we evaluate the procedures that would be necessary to embed such a framework into clinical decision making to contribute to the prospective identification of therapy targets most likely to trigger an immune-mediated abscopal response. We additionally highlight areas in which caution is required when integrating the tools of mathematical oncology into the clinic.
2. Feasibility of clinical application
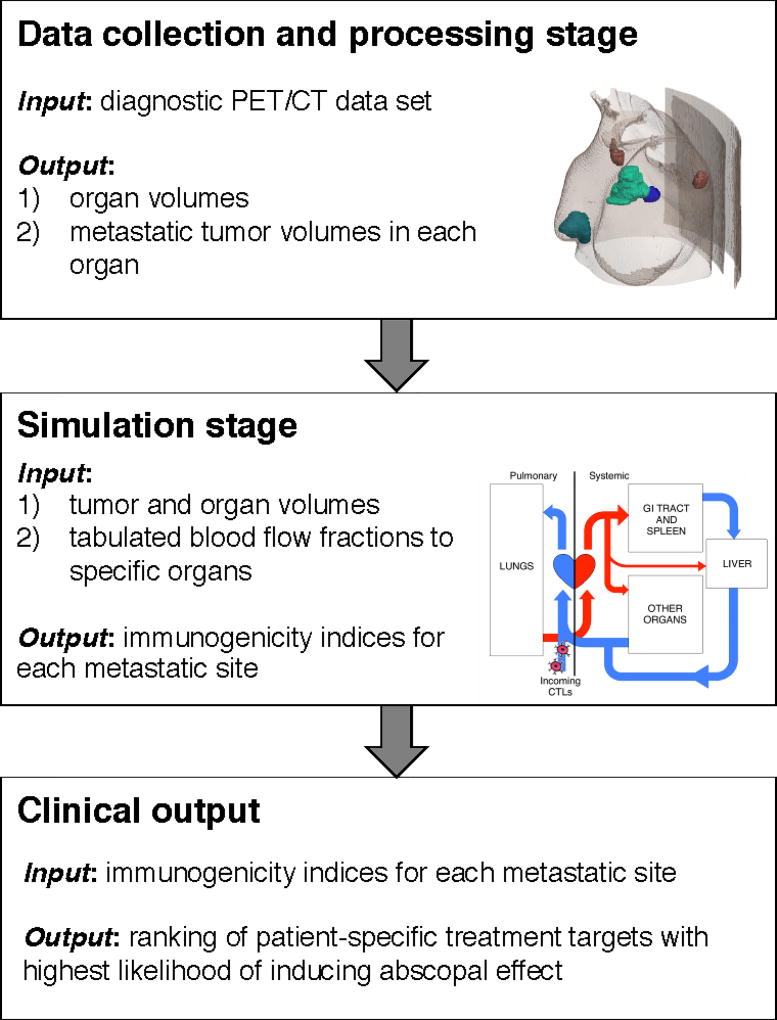
There would be three independent stages involved in the patient-specific clinical application of the proposed T cell trafficking framework: 1) data collection and processing; 2) computer simulations; and 3) clinical recommendations (see Fig. 1). During the data collection and processing stage, tumor volumes in each respective organ would need to be obtained from clinical imaging. With this patient-specific information and corresponding physiologic blood flow fraction to each tumor bearing organ, the calibrated model could be initialized and T cell trafficking between respective metastatic sites simulated. Based on model predictions of T cell dissemination patterns, the immunogenicity index of each tumor site could be calculated. The metastatic site with the highest immunogenicity index would then be identified from ranking these indices, and determined to have the greatest potential for inducing an anti-tumor abscopal effect. This treatment target recommendation can be provided via the computational tool to the attending physician to aid decision making at their discretion.
Figure 1.
Clinical workflow of the proposed T cell trafficking framework.
2.1. Data collection and processing stage
To initialize the model, patient-specific data in the form of PET/CT or MRI scans covering all or many metastatic sites need to be collected. CT scans of the chest, abdomen and pelvis are recommended by clinical guidelines in the staging of, amongst others, metastatic colon/rectal and breast cancers [9, 10]. This means the data necessary to initialize the model will typically already be collected and available in the clinic.
Imaging data needs to be further processed in order to extract more detailed information about volumes of tumor-bearing organs and existing metastatic sites [5]. In current clinical practice, only the maximal diameters of the tumors of interest are recorded to evaluate treatment response [11], and 3D tumor volumes are only delineated for radiation treatment planning purposes [12]. Furthermore, the latter is only being conducted for one site that has been chosen to be targeted by radiotherapy. To utilize the model one would need to delineate the 3D volume of all existing sites, which in a heavily metastatic setting may be unrealistic; manual delineation of, for example, an isolated lung tumor can take on average around 16 minutes for a qualified medical practitioner [13]. Delineation time can be reduced, however, by using a PET scan to provide a software-generated contour for further editing by the observer [13, 14].
The proposed T cell trafficking framework also requires information relating to the volume of tumor-bearing organs. Delineation of anatomical structures is less challenging, however, thanks to a priori knowledge of the 3-dimensional shapes of specific organs. Atlas- and shape-based segmentation tools use reference image volumes (the atlas) that have already been segmented to perform constrained segmentation of individual structures [15]. To reduce the risk of error, delineation obtained from automated segmentation could be further adjusted by the attending radiologist. Furthermore, as only the volume and not exact delineation is needed, one could measure only maximal diameters of the organ in all 3 planes and scale them to the known organ shape stored in the anatomical atlas. This could minimize the time required for obtaining organ volumes.
2.2. Simulation stage
Despite the inherent challenges of collecting tumor distribution and volume data, once obtained, these patient-specific volume measurements are the primary input to the mathematical model. Also crucial for patient-specific simulations are blood flow fractions (BFFs, % of cardiac output) to each tumor-bearing organ. Average values of these BFFs have been measured and tabulated for major organs in healthy individuals [16] and can be used as a first approximation in the model. In the case of organs for which BFFs have not been established, one could estimate their values using one of several mathematical models of blood flow through the arterial tree that have been extensively developed during the last several decades [17, 18, 19, 20]. In a recent publication by Blanco and colleagues [19] the architecture of the blood flow model comprises almost every arterial vessel acknowledged in the medical/anatomical literature, with a resolution down to the luminal area of perforator arteries.
With both tumor/organ volumes and respective blood flow fractions, the probability of T cell infiltration of a particular tumor can be calculated for each metastatic site, and the entropy of final dissemination of T cells can guide the calculation of each respective site’s immunogenicity index. The metastatic site-specific immunogenicity index takes into account both the relative metastatic tumor volume, and the entropy of final T cell distribution among all metastatic sites [5]. The relative metastatic tumor volume is expressed as the ratio of metastatic site volume to the volume of the largest measured tumor, and reflects the assumption that the larger the tumor, the more APCs will be activated. The inclusion of the entropy of final T cell distribution reflects the requirement of the most uniform spread of T cells possible. This entropy is expressed as the ratio of the metastatic site-specific T cell distribution entropy to the entropy of the uniform distribution.
Model simulations run in a very short time, and can calculate immunogenicity indices of metastatic tumor sites almost instantaneously. The computational tool itself is highly amenable to clinical application due to this high computational efficiency.
2.3. Clinical output
The primary output of the model, i.e. immunogenicity indices for each metastatic site, could be ranked and presented to the attending physician. Poleszczuk and colleagues hypothesized in [5] that the site with the highest immunogenicity index should be the primary radiotherapy target, as radiation applied to that site may elicit the highest systemic (abscopal) response.
It should be noted that in some instances the clinician-recommended treatment site may be selected for necessity, i.e. for palliative care to relieve symptoms as opposed to with curative intent. In this case, where possible, a secondary treatment site would need to be considered. In this case, the proposed framework could be extended to calculate immunogenicity indices for two or more treatment sites to irradiate, without jeopardizing computational speed.
3. Clinical trial design
Any clinical decision support system needs to be thoroughly verified in a prospective clinical trial prior to application in the clinic. Validation of the findings of Poleszczuk and colleagues would require the development of a clinical trial testing the hypothesis that irradiating sites identified by model-calculated immunogenicity indices leads to an increased systemic (abscopal) response when compared to unguided target selection. Prospective clinical trials evaluating abscopal response after irradiation are, however, scarce, with only one from which results have been published to our knowledge [21]. Thus, designing a clinical trial protocol taking into account all possible confounding factors is a complicated task.
One can envision that in such a clinical trial all enrolled patients will have previously demonstrated symptomatic or radiologic disease progression, with no sustained clinical benefit from prior therapy. An additional criteria for recruitment would be the physical ability of the patient to undergo PET/CT scans of at least chest, abdomen and pelvis. In addition, it may be necessary to limit the maximal number of existing metastatic sites due to the time constraints associated with tumor delineation highlighted in the previous section.
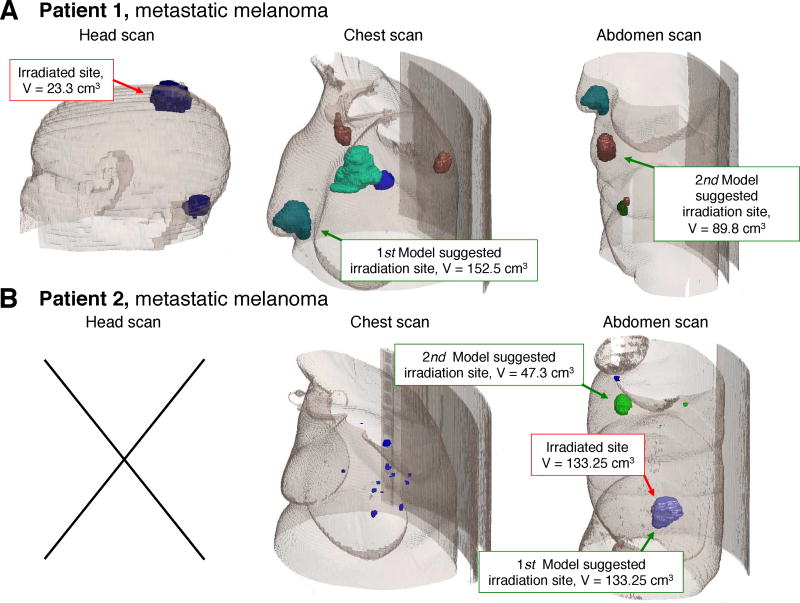
Two potential study arms would be required; cohort A in which target site is determined by the clinician, and cohort B in which target site is determined by the model. This study design depends on the assumption that model-recommended sites will differ from clinician-recommended sites in a sufficient number of cases to recruit the required sample size. Although limited data exists to verify this assumption at present, from two existing patients for which simulations of T cell redistribution between the predefined metastatic sites have been obtained, the actual treatment site differs from the model-predicted site in one instance (Fig. 2). To further address this issue, one could consider a Simons optimal two-stage design for this trial; an additional group of patients would be enrolled only if the model predicted different treatment targets to the clinician in at least one patient of the first N. If no difference in predicted sites were to be observed in the first group of patients, the trial would be terminated.
Figure 2.
Model-suggested target sites as compared to actual treatment sites in two independent metastatic melanoma patients.
In the model guided arm of the clinical trial, cases in which irradiation of the model-predicted metastatic site is infeasible due to, for example, anatomical location having too high constraints for normal tissue toxicities must also be considered. In this case, the first feasible irradiation site of the sites ranked by immunogenicity index could be selected.
To fully harness the synergy of radiation and the immune system, it may be sufficient to deliver limited immune-priming radiation (35 Gy in 10 fractions over 2 weeks [21], or 24 Gy in 3 fractions [22]) to the patients in both arms of the trial. In order to increase the chance of observing a significant abscopal response, radiation could be enhanced with concurrent/subsequent immunotherapy such as an anti-CTLA4 antibody [23], anti-programmed death 1 antibody [24], or granulocyte-macrophage colony-stimulating factor [21].
3.1. Evaluating outcomes
Should the model-informed patients demonstrate a higher response rate defined by the above criteria, we may conclude that optimizing redistribution of activated T cells may in fact increase the likelihood of systemic responses and a greater reduction in tumor burden.
Initially, treatment success and failure need to be clearly defined. In existing clinical trials, a responder has been defined as a patient for whom at least a 30% decrease in the longest diameter of any measurable (1 cm) non-irradiated lesion from baseline was observed [21]. The framework proposed by Poleszczuk and colleagues aims, however, at increasing the systemic response in terms in the overall tumor burden. Therefore, it seems more appropriate to assess the overall tumor burden reduction in the patient, excluding the volume of the irradiated tumor.
An additional shortcoming of existing trial design is that typically the observation of only one growing tumor is required for classifying a patient as a non-responder to prior treatment. It is conceivable that patients enrolled in a second trial failed previous therapy by this classification, but had one or more metastatic sites responding to the prior treatment. This has been observed in clinical trials testing adoptive T cell transfer [25]. Thus, growth patterns of each metastatic site prior to treatment should also be evaluated to ensure any sites where the tumor was already decreasing in volume are excluded from subsequent post-therapy evaluation, and are not a confounding factor in quantifying the abscopal effect. This information could be obtained from previous clinical intervention, if applicable.
4. Discussion
To guide robust and validatable mathematical models, the identification of mechanisms for which clinical and experimental data is readily available is crucial. Attempting to elucidate mechanisms governing patient response to treatment based on a vast wealth of biological knowledge but only a limited amount of clinically obtainable patient information is a near-impossible task. By isolating individual mechanisms that contribute to the larger, more complex biological scenario, such as the role of activated T cell dissemination and systemic immune responses in reduction of overall tumor burden, more tractable methods can be used to answer specific, focused questions. Where data-driven mathematical models suggest the role of these mechanisms may be substantial, the mechanism in question can be assessed for its ability to aid decision making in the clinical setting.
The T cell trafficking model proposed by Poleszczuk and colleagues [5] provides suggestions of the optimal site(s) for irradiation in order to increase the magnitude of immune-mediated abscopal responses. This model is partially calibrated with experimental and clinical data, requires only patient-specific tumor/organ volume data at each metastatic site to initiate simulations and generate predictions, and thus has the potential to aid clinical decision making in the future. That being said, further crucial steps must be taken to prepare such a tool for application in the clinical setting. Initially, this includes the verification of all physiologic assumptions such as the pre-calculated blood flow fractions to specific organs for biological accuracy.
Although current imaging techniques including MRI and PET/CT can identify the distribution of metastatic sites in an individual patient, methods for the efficient delineation of such scans in the case of multiple metastasis need to be established for timely collection and analysis of patient data. Known inaccuracies in tumor delineation techniques could also affect model predictions [14]. Furthermore, a detailed evaluation of the ability of radiation of the suggested target sites to induce the hypothesized abscopal response needs to be conducted. This requires a particularly careful clinical trial design to isolate abscopal responses from other confounding factors and demonstrate the superiority of model identified targets to those typically recommended in the clinical setting.
An important thing to note when considering the integration of the tools of mathematical oncology into the clinic is that these tools often feature heavily simplified models representing significantly more complex and dynamic biological systems. Such tools do not seek or profess to capture all scales or components of complex and dynamic biological systems, but rather aspire to elucidate and understand individual mechanisms and, if possible, use this understanding to contribute to the improvement of patient outcomes. Many additional factors could have been considered in the model proposed by Poleszczuk and colleagues [5]. After redistribution by the circulatory system, the ability of activated T cells to proliferate and expand at each respective metastatic site is likely to contribute to the magnitude of the abscopal response observed in that site. Moreover, the local inflammatory environment at each respective site and the quality of tumor neovasculature which is often fragile and prone to leakage may alter T cell extravasation rates, especially at the treated tumor site [26, 27]. As non-invasive identification of microenvironmental conditions and correlation with T cell extravasation or expansion is currently impossible, these factors were excluded from the original model. Radiomics habitat analysis may hold promise to provide such biomarkers in the future [28, 29, 30], yet this may take many more years of development and validation. Thusly, due to the large degree of simplification, the proposed model and all such models require extremely thorough validation in the clinical setting and should be utilized only as a complement to the expertise and experience of the attending clinician.
Acknowledgments
This work is supported in part by the Personalized Medicine Award 09-33000-15-03 from the DeBartolo Family Personalized Medicine Institute Pilot Research Awards in Personalized Medicine (PRAPM), and by the the Moffitt Cancer Center PSOC, NIH/NCI U54CA143970 and U54CA193489.
References
- 1.Coussens Lisa M, Werb Zena. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.González Fermín E, Gleisner Alejandra, Falcón-Beas Felipe, Osorio Fabiola, López Mercedes N, Salazar-Onfray Flavio. Tumor cell lysates as immunogenic sources for cancer vaccine design. Human vaccines & immunotherapeutics. 2014;10(11):3261–3269. doi: 10.4161/21645515.2014.982996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reits Eric A, Hodge James W, Herberts Carla A, Groothuis Tom A, Chakraborty Mala, Wansley Elizabeth K, Camphausen Kevin, Luiten Rosalie M, de Ru Arnold H, Neijssen Joost, et al. Radiation modulates the peptide repertoire, enhances mhc class i expression, and induces successful antitumor immunotherapy. The Journal of experimental medicine. 2006;203(5):1259–1271. doi: 10.1084/jem.20052494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vatner Ralph E, Cooper Benjamin T, Vanpouille-Box Claire, Demaria Sandra, Formenti Silvia C. Combinations of immunotherapy and radiation in cancer therapy. Frontiers in oncology. 2014;4:325. doi: 10.3389/fonc.2014.00325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poleszczuk Jan T, Luddy Kimberly A, Prokopiou Sotiris, et al. Abscopal benefits of localized radiotherapy depend on activated t-cell trafficking and distribution between metastatic lesions. Cancer Res. 2016;76(5):1009–1018. doi: 10.1158/0008-5472.CAN-15-1423. [DOI] [PubMed] [Google Scholar]
- 6.Rockne Russel, Rockhill JK, Mrugala M, et al. Predicting the efficacy of radiotherapy in individual glioblastoma patients in vivo: a mathematical modeling approach. Phys Med Biol. 2010;55(12):3271. doi: 10.1088/0031-9155/55/12/001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prokopiou Sotiris, Moros Eduardo G, Poleszczuk Jan, et al. A proliferation saturation index to predict radiation response and personalize radiotherapy fractionation. Radiat Oncol. 2015;10(1):1. doi: 10.1186/s13014-015-0465-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yankeelov Thomas E, Atuegwu Nkiruka, Hormuth David, et al. Clinically relevant modeling of tumor growth and treatment response. Sci Transl Med. 2013;5(187):187ps9. doi: 10.1126/scitranslmed.3005686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith Travis J, Korngold Elena, Orloff Susan L. Preoperative imaging in colorectal liver metastases: current practices. Current Surgery Reports. 2014;2(2):1–9. [Google Scholar]
- 10.Gold Laura S, Lee Christoph I, Devine Beth, Nelson Heidi, Chou Roger, Ramsey Scott, Sullivan Sean D. Imaging techniques for treatment evaluation for metastatic breast cancer. 2014 [PubMed] [Google Scholar]
- 11.Eisenhauer EA1, Therasse Patrick, Bogaerts Jan, Schwartz LH, Sargent D, Ford Robert, Dancey J, Arbuck S, Gwyther S, Mooney M, et al. New response evaluation criteria in solid tumours: revised recist guideline (version 1.1) European journal of cancer. 2009;45(2):228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 12.Njeh CF, et al. Tumor delineation: The weakest link in the search for accuracy in radiotherapy. Journal of Medical Physics. 2008;33(4):136. doi: 10.4103/0971-6203.44472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steenbakkers Roel JHM, Duppen Joop C, Fitton Isabelle, Deurloo Kirsten EI, Zijp Lambert J, Comans Emile FI, Uitterhoeve Apollonia LJ, Rodrigus Patrick TR, Kramer Gijsbert WP, Bussink Johan, et al. Reduction of observer variation using matched ct-pet for lung cancer delineation: a three-dimensional analysis. International Journal of Radiation Oncology* Biology* Physics. 2006;64(2):435–448. doi: 10.1016/j.ijrobp.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 14.Van Baardwijk Angela, Bosmans Geert, Boersma Liesbeth, Buijsen Jeroen, Wanders Stofferinus, Hochstenbag Monique, Van Suylen Robert-Jan, Dekker André, Dehing-Oberije Cary, Houben Ruud, et al. Pet-ct–based auto-contouring in non–small-cell lung cancer correlates with pathology and reduces interobserver variability in the delineation of the primary tumor and involved nodal volumes. International Journal of Radiation Oncology* Biology* Physics. 2007;68(3):771–778. doi: 10.1016/j.ijrobp.2006.12.067. [DOI] [PubMed] [Google Scholar]
- 15.Whitfield Gillian A, Price P, Price Gareth J, Moore Christopher J. Automated delineation of radiotherapy volumes: are we going in the right direction? The British journal of radiology. 2013;86(1021):20110718–20110718. doi: 10.1259/bjr.20110718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valentin Jack. Basic anatomical and physiological data for use in radiological protection: reference values: Icrp publication 89. Annals of the ICRP. 2002;32(3):1–277. [PubMed] [Google Scholar]
- 17.Reymond Philippe, Merenda Fabrice, Perren Fabienne, Rüfenacht Daniel, Stergiopulos Nikos. Validation of a one-dimensional model of the systemic arterial tree. American Journal of Physiology-Heart and Circulatory Physiology. 2009;297(1):H208–H222. doi: 10.1152/ajpheart.00037.2009. [DOI] [PubMed] [Google Scholar]
- 18.van de Vosse Frans N, Stergiopulos Nikos. Pulse wave propagation in the arterial tree. Annual Review of Fluid Mechanics. 2011;43:467–499. [Google Scholar]
- 19.Blanco Pablo J, Watanabe Sansuke M, Passos Marco Aurélio RF, Lemos Pedro A, Feijóo Raúl A. An anatomically detailed arterial network model for one-dimensional computational hemodynamics. IEEE Transactions on Biomedical Engineering. 2015;62(2):736–753. doi: 10.1109/TBME.2014.2364522. [DOI] [PubMed] [Google Scholar]
- 20.Olufsen Mette S, Peskin Charles S, Kim Won Yong, Pedersen Erik M, Nadim Ali, Larsen Jesper. Numerical simulation and experimental validation of blood flow in arteries with structured-tree outflow conditions. Annals of biomedical engineering. 2000;28(11):1281–1299. doi: 10.1114/1.1326031. [DOI] [PubMed] [Google Scholar]
- 21.Golden EB, Demaria S, Schiff PB, et al. An abscopal response to radiation and ipilimumab in a patient with metastatic non-small cell lung cancer. Cancer Immunol Res. 2013;1(6):365–372. doi: 10.1158/2326-6066.CIR-13-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dewan M Zahidunnabi, Galloway Ashley E, Kawashima Noriko, et al. Fractionated but not single-dose radiotherapy induces an immune-mediated abscopal effect when combined with anti–ctla-4 antibody. Clin Cancer Res. 2009;15(17):5379–5388. doi: 10.1158/1078-0432.CCR-09-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolchok Jedd D, Saenger Yvonne. The mechanism of anti-ctla-4 activity and the negative regulation of t-cell activation. The Oncologist. 2008;13(Supplement 4):2–9. doi: 10.1634/theoncologist.13-S4-2. [DOI] [PubMed] [Google Scholar]
- 24.Park SS, Dong H, Liu X, Harrington SM, Krco CJ, Grams MP, Mansfield AS, Furutani KM, Olivier KR, Kwon ED. Pd-1 restrains radiotherapy-induced abscopal effect. Cancer Immunol Res. 2015;3(6):610–9. doi: 10.1158/2326-6066.CIR-14-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilon-Thomas Shari, Kuhn Lisa, Ellwanger Sabine, Janssen William, Royster Erica, Marzban Suroosh, Kudchadkar Ragini, Zager Jonathan, Gibney Geoffrey, Sondak Vernon K, et al. Brief communication: Efficacy of adoptive cell transfer of tumor infiltrating lymphocytes after lymphopenia induction for metastatic melanoma. Journal of immunotherapy (Hagerstown, Md.: 1997) 2012;35(8):615. doi: 10.1097/CJI.0b013e31826e8f5f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melero I, Rouzaut A, Motz GT, Coukos G. T-cell and nk-cell infiltration into solid tumors: a key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014;4(5):522–526. doi: 10.1158/2159-8290.CD-13-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lanitis Evripidis, Irving Melita, Coukos George. Targeting the tumor vasculature to enhance t cell activity. Curr Opin Immunol. 2015;33:5563. doi: 10.1016/j.coi.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aerts HJ. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2016;2(12):1636–1642. doi: 10.1001/jamaoncol.2016.2631. [DOI] [PubMed] [Google Scholar]
- 30.Lee G, Lee HY, Park H, Schiebler ML, van Beek EJ, Ohno Y, Seo JB, Leung A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur J Radiol. 2017;86:297–307. doi: 10.1016/j.ejrad.2016.09.005. [DOI] [PubMed] [Google Scholar]