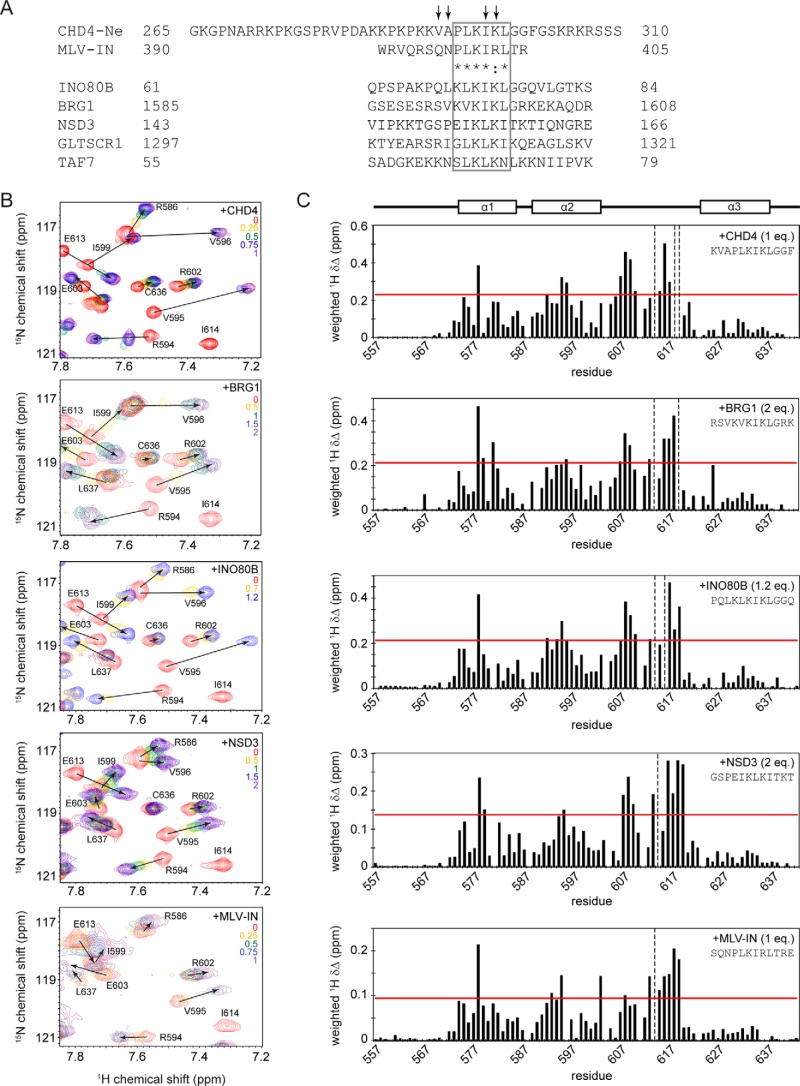
Figure 4.
BRD3-ET can interact in a conserved manner with motifs from a range of transcriptional coregulators. A, sequence alignment of CHD-Ne with a peptide from MLV-IN that is known to bind the ET domain of Brd4 (50), as well as related sequences found in INO80B, BRG1, NSD3, GLTSCR1, and TAF7. A region of high similarity (representing a potential ET recognition motif) is boxed. Residues that display the largest chemical shift perturbations in the presence of BRD3-ET are indicated by arrows. B, partial 15N HSQC spectra of BRD3-ET alone (red) and titrated with peptides from (top to bottom) CHD4, BRG1, INO80B, NSD3, and MLV-IN. The number of molar equivalents of each peptide added are indicated in each spectrum. C, chemical shift changes induced in BRD3 ET domain at saturation with CHD4, INO80B, and NSD3. The horizontal red line indicates 1 S.D. above mean chemical shift change. Peaks no longer visible at the titration end point are indicated by vertical dashed lines. Residues that are identical or highly similar between CHD4 and MLV integrase are indicated by asterisks and a colon, respectively.