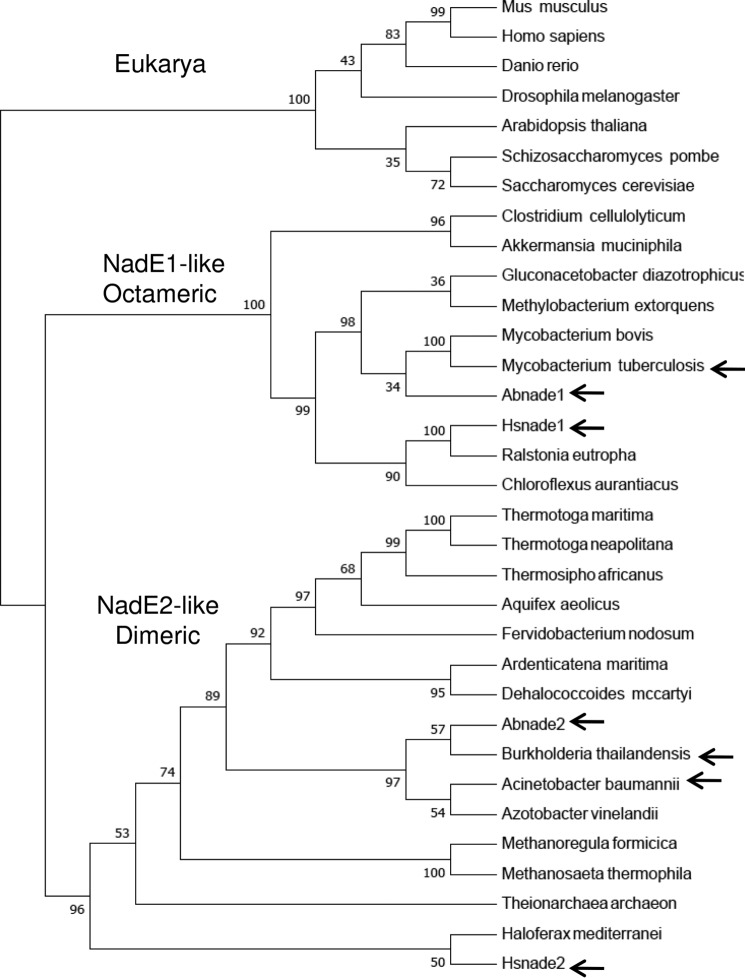
Figure 4.
Phylogenetic analysis of selected NadEGln. The sequence of NadEGln was retrieved from NCBI and aligned using Clustal W. The phylogentic three was constructed by Neighbor-joining using MEGA 7. All positions containing gaps and missing data were eliminated from the data set. Bootstrap values were adjusted to 1000 replicates. Three groups were observed: 1) eukaryotic representatives; 2) NadE1-like octameric NadEGln; and 3) NadE2-like dimeric NadEGln. The relevant proteins with experimentally determined quaternary structure are indicated by arrows. A. baumannii, WP_065718975.1; Akkermansia muciniphila ATCC BAA-835, ACD04457.1; Aquifex aeolicus, NP_213654.1; Arabidopsis thaliana, NP_175906.1; Ardenticatena maritima, KPL88222.1; A. brasilense, AIB10872.1; A. brasilense, AIB14429.1; Azotobacter vinelandii DJ, YP_002798395.1; B. thailandensis, AOJ56104.1; Chloroflexus aurantiacus, ABY34602.1; Clostridium cellulolyticum H10, YP_002505537.1; Danio rerio, NP_001092723.1; Dehalococcoides mccartyi 195, YP_181837.1; Drosophila melanogaster, NP_572913.1; Fervidobacterium nodosum Rt17-B1, YP_001410277.1; Gluconacetobacter diazotrophicus PA1 5, YP_001601200.1; Haloferax mediterranei, AHZ23047.1; H. seropedicae, AKN65438.1; H. seropedicae, AKN67808.1; Homo sapiens, NP_060631.2; Methanoregula formicica, AGB01500.1; Methanosaeta thermophila PT, YP_843157.1; Methylobacterium extorquens CM4, YP_002423109.1; Mus musculus, NP_084497.1; Mycobacterium bovis AF2122/97, NP_856111.1; Ralstonia eutropha H16, YP_725265.1; S. cerevisiae S288C, NP_011941.1; Schizosaccharomyces pombe 972h-, NP_587771.1; Theionarchaea archaeon, KYK35595.1; Thermosipho africanus TCF52B, YP_002335497.1; T. maritima, AKE27162.1; Thermotoga neapolitana DSM 4359, YP_002534863.1; and M. tuberculosis, AMP30329.1.