Figure 1.
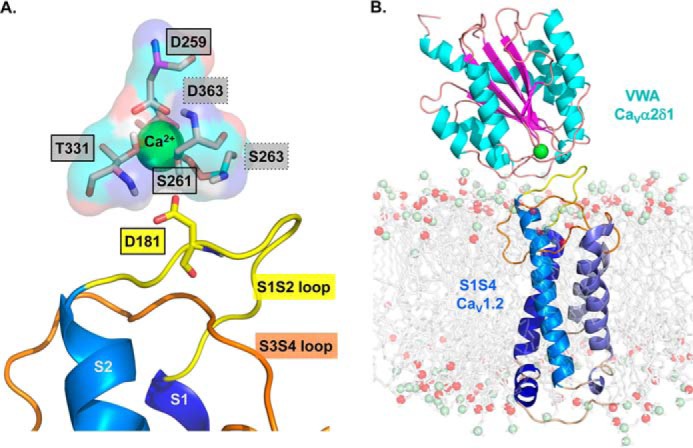
3D homology model of the S1 to S4 region from domain I of CaV1.2 in complex with the VWA domain of CaVα2δ1. A homology model of the CaV1.2–CaVα2δ1 interface was built based using the molecular coordinates of the cryo-EM structure of the skeletal muscle CaV1.1 complex (PDB code 5GJV). This model differs slightly from the 3D model of the same region published previously (19) in regard to the orientation of the IS3S4 extracellular loop. A, residues forming the protein interface are zoomed in with emphasis on the MIDAS motif. The main-chain groups of amino acids forming the MIDAS motif (Asp-259, Ser-261, Ser-263, Thr-331, and Asp-363) are shown as pale rose sticks with carboxyl groups in red, amine groups are blue, and hydrogen atoms are white. Residues Asp-259, Ser-261, and Thr-331 identified within full-lined black boxes are in the foreground, and Ser-263 and Asp-363 identified within the dot-lined boxes are in the background. MIDAS residues are facing the IS1S2 loop in CaV1.2. CaV1.2 Asp-181 is shown in stick representation. B, VWA domain of the rat CaVα2δ1 is shown in schematic representation in which α-helices appear in cyan, and β-strands are shown in pink. A single Ca2+ ion (green) is shown as being coordinated by the MIDAS residues. The region spanning from the S1 to the S4 segments in repeat I (IS1S4) of CaV1.2 is shown in schematic representation, and the transmembrane helices S1 to S4 are colored from the darker (S1) to the paler shade of blue (S4). Modeled DPPC lipids are shown in stick representation with carbon, oxygen, and phosphorus atoms being shown in gray, red, and pale green, respectively. Modeling was achieved with Modeler 9.17. The figure was produced using PyMOL (DeLano Scientific).
