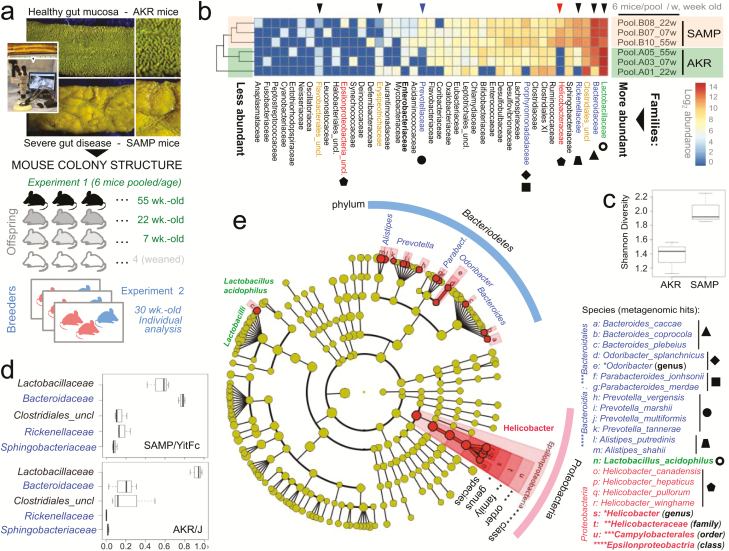
FIGURE 1.
Metagenomic analysis of pooled fecal matter reveals an abundance of Bacteroidetes and Helicobacteraceae in SAMP mice. A, En face stereomicroscopic images of healthy AKR ileal mucosa (top image) and SAMP cobblestone ileitis (bottom image); schematic illustration of metagenomic sampling design in SPF facilities (pooled feces in experiment 1 [summer] vs individual sampling in experiment 2 [8 months later, spring]). B, Multivariable unsupervised hierarchical analysis of metagenomic fecal bacterial abundance from 36 mice pooled as 6 mice per age group (7, 22, and 55 weeks old). Note clustering of AKR and SAMP samples as separate clades. Arrows illustrate distinct families, ranked by abundance across all samples. C, Compared with AKR, the SAMP metagenome have greater bacterial diversity (Shannon diversity). D, Boxplots reveal reduction of Firmicutes (Lactobacillaceae and Clostridiales) and expansion of Bacteroidetes families in SAMP (eg, Bacteroidaceae, Rickenellaceae, Sphingobacteriaceae). E, Circular diagram of relative taxonomic metagenomic abundance in phylogenetic tree format highlights significant enrichment of Helicobacteraceae (Proteobacteria phylum), and 4 of 6 known Bacteroidetes phylum families (Bacteroidaceae, Rickenellaceae, Porphyromonadaceae, and Prevotellaceae) in SAMP mice (1-tailed Fisher exact P = 0.046).