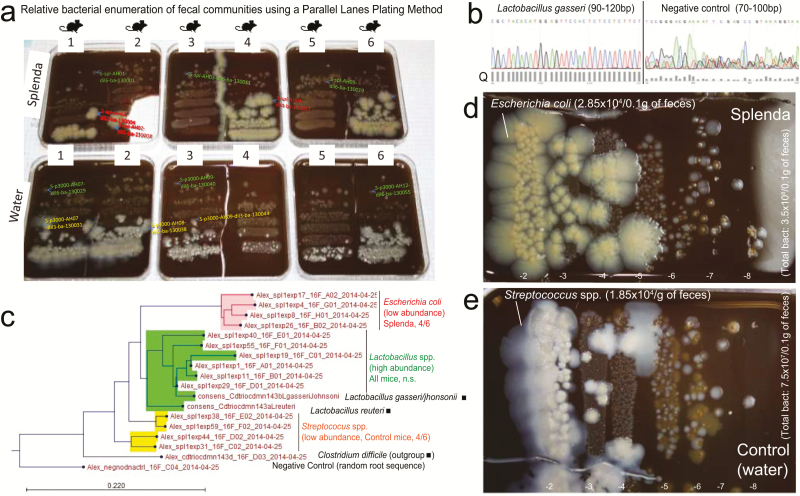
FIGURE 6.
Splenda promotes the replacement of Streptococcus spp. with E. coli in the intestinal tract of SAMP mice. A, “Parallel Lanes Plating” Method. Photograph of BHI agar plates supplemented with 5% sheep defibrinated blood inoculated with 10-fold serial dilutions of feces from SAMP mice using a parallel minidrop slide and lanes method developed for tracking and relative enumeration of complex bacterial communities (Supplementary Fig. 1). Mice were caged individually and exposed to a composite of bedding and feces (IsPreFeH) prior to receiving Splenda at a low dose (1.5 mg/mL) or water for 42 days. Representative colonies comprising all possible morphologies in each agar plate were selected for purification and Sanger sequencing for speciation (from high dilutions, green labels; and low dilutions, pink or red labels). Notice the colony morphology (large, thick, spreading) of 4/6 mice in the Splenda group is different from that of 4/6 control mice (whitish, smaller). B, Sanger sequence chromatograph. Single-colony PCR revealed Lactobacillus gasseri (umbonate, brown) as the most common abundant bacteria in mice, unaffected by Splenda supplementation (see the panels below). Q, quality of consensus sequence. C, Phylogenetic analysis of 16S rRNA paired-end consensus sequences revealed that the whitish colonies in the control (water) group were closely related to Streptococcus spp., while bacteria in Splenda mice were E. coli. D and E, Close-up of colony morphologies on BHI agar after 5 days of aerobic incubation. Notice the “parallel lanes method” of 2 mice representing the Splenda and control groups. Negative numbers indicate 10-fold dilution factor.