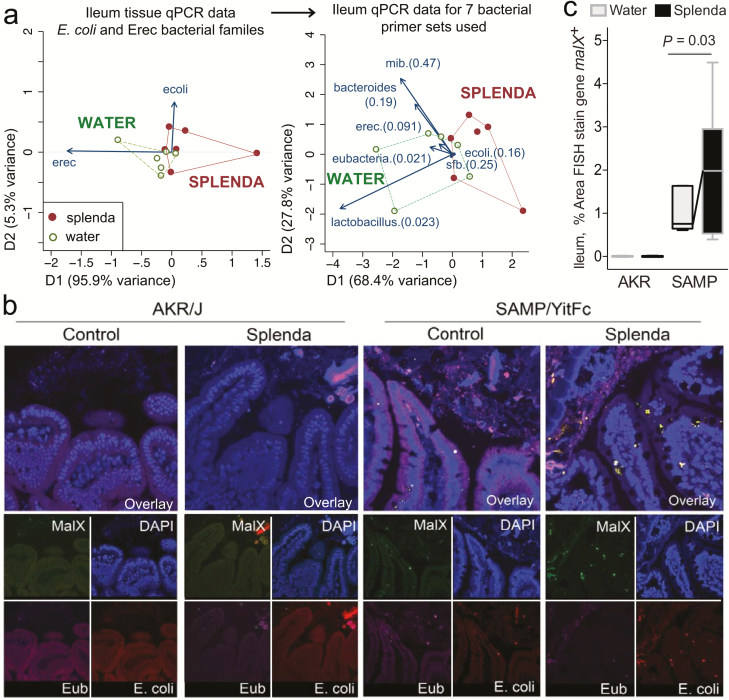
FIGURE 7.
Bacterial qPCR and FISH staining of ileal tissue illustrates distinct microbiota and increased invasive malX+ bacteria (E. coli) in Splenda-supplemented SAMP but not AKR mice. A, Multivariate analysis of DNA qPCR data from ileum tissues of SAMP mice after 42 days of supplementation. Notice the display of mice (points) and the vector influence of the variables (arrows) on the overall matrix data variability (D1 and D2) for the E. coli and Erec primers, and for all the 7 primer sets used in this study. Hotelling’s T-test P values are in parentheses. Notice the separation of the 2 clusters (water, Splenda). B, Ileal sections from SAMP mice supplemented with 3.5% Splenda for 42 days (Splenda) or nontreated control mice (Water) were hybridized with probes to Eubacteria (purple), E. coli (red), and malX (maldodextrin, green), a component of the maltose/maltodextrin metabolism system. Cell nuclei are visualized with DAPI (blue). Images shown are representative of analyses performed in 5 mice per group. Notice the presence of E. coli in both the epithelial layer and the subepithelial lamina propria tissue (villi), and the large bacterial clusters in the lamina propria of SAMP mice on the Splenda panels. C, Percentage of area that stained positive for the malX gene probe (pixels, malX+ area/total tissue area*100). Unpaired t test statistics. A minimum of 4 fields were analyzed/sampled using ImageProPlus v7 software (AKR control, n = 4; AKR Splenda, n = 6; SAMP control, n = 6; SAMP Splenda, n = 10).