Abstract
The differentiation status of neuroblastoma (NB) strongly correlates with its clinical outcomes; however, the molecular mechanisms driving maintenance of stemness and differentiation remain poorly understood. Here, we show that plant homeodomain finger-containing protein 20 (PHF20) functions as a critical epigenetic regulator in sustaining stem cell-like phenotype of NB by using CRISPR/Cas9-based targeted knockout (KO) for high-throughput screening of gene function in NB cell differentiation. The expression of PHF20 in NB was significantly associated with high aggressiveness of the tumor and poor outcomes for NB patients. Deletion of PHF20 inhibited NB cell proliferation, invasive migration, and stem cell-like traits. Mechanistically, PHF20 interacts with poly(ADP-ribose) polymerase 1 (PARP1) and directly binds to promoter regions of octamer-binding transcription factor 4 (OCT4) and sex determining region Y-box 2 (SOX2) to modulate a histone mark associated with active transcription, trimethylation of lysine 4 on histone H3 protein subunit (H3K4me3). Overexpression of OCT4 and SOX2 restored growth and progression of PHF20 KO tumor cells. Consistently, OCT4 and SOX2 protein levels in clinical NB specimens were positively correlated with PHF20 expression. Our results establish PHF20 as a key driver of NB stem cell-like properties and aggressive behaviors, with implications for prognosis and therapy.
Keywords: PHF20, neuroblastoma, PARP1, cancer stem cell-like traits, epigenetic regulation
Introduction
Neuroblastoma (NB) is the most common extracranial solid tumor of childhood, accounting for the largest number of cancer-related deaths in children (Louis and Shohet, 2015). This tumor arises from the developing neural crest cells, which possess self-renewal and multipotency characteristics (Huang and Weiss, 2013), and aberrations in normal developmental processes are most likely its primary cause (Takahashi et al., 2013). Notably, patients with undifferentiated or poorly differentiated NB have significantly worse outcomes than those with well-differentiated NB (Molenaar et al., 2012). Therefore, the identification of key regulators that control NB risk stratification is critically important for developing more effective therapeutics.
Several markers that predict a good or poor treatment outcome have been reported (Janoueix-Lerosey et al., 2009). Currently, the most validated prognostic marker of high-risk disease and poor prognosis is amplification of the NB-derived v-myc avian (MYCN) oncogene (Powers et al., 2016), which is present in ~25% of cases (Huang and Weiss, 2013). MYCN is involved in the regulation of self-renewal and can substitute for MYC in reprogramming fibroblasts into induced pluripotent stem cells (iPSC) (Smith et al., 2010). Emerging evidence from several tumor types, including NB, points to the potential active role of the cancer initiating cells in disease progression, relapse, and poor outcomes (Suva et al., 2013). Thus, MYCN drives NB into a stem cell-like state by blockade of differentiation pathways and expression of self-renewal and pluripotency factors (Kaneko et al., 2015).
Some NB cells retain multipotency and overexpress stem cell-related genes, such as OCT4 and SOX2 (Singovski et al., 2016). OCT4, SOX2, and NANOG have been demonstrated to play critical roles in stem cell self-renewal and have been proposed to promote the self-renewal of cancer cells with stem cell-like properties (Mu et al., 2017). Despite these correlative studies between OCT4 and SOX2 expression and the stem cell-like state of NB, how OCT4 and SOX2 are reactivated for conferring NB stem cell-like traits remains unclear.
Systematic search and analysis for genomic alterations using whole-genome or whole-exome sequencing show that NB has remarkably low genetic complexity along with few genes that have significant mutations (Chmielecki et al., 2017). These findings indicate that aberrant epigenetic modifications, including DNA methylation and histone modification (Olsson et al., 2016), are important features of both development and progression of NB. However, their functional relevance is largely unknown. In this study, we developed a targeted knockout (KO) strategy and conducted a screening of 573 transcriptional and epigenetic factors required for NB differentiation. Among the genes identified, we found that plant homeodomain finger-containing protein 20 (PHF20) was a key epigenetic factor controlling the stem cell-like phenotype of NB. Ablation of PHF20 led to inhibition of proliferation and malignancy, while ectopic expression of PHF20 enhanced the expression of OCT4 and SOX2, suggesting that PHF20 is a pivotal regulator of NB initiation and progression. Thus, our findings have identified PHF20 as a therapeutic target for NB treatment.
Results
Identification of PHF20 as a driver of stem cell-like phenotype in NB
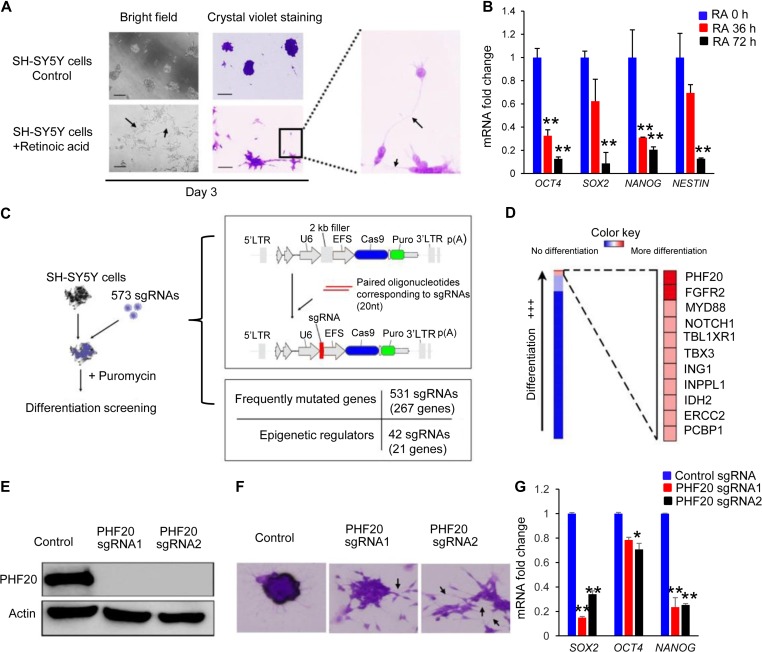
Identification of key factors that regulate cancer initiation and progression may help develop novel and effective strategies to overcome the chemoresistance associated with NB therapy. Thus, we designed a high-throughput screening based on a CRISPR/Cas9 library of 573 sgRNAs to identify potential targets (Supplementary Table S1). This screening targeted 288 genes, which included frequently mutated genes and epigenetic regulators. As shown in Figure 1A, retinoic acid (RA)-treated SH-SY5Y cells showed intense neurite networking by Day 3, while untreated SH-SY5Y cells formed aggregates over time. Stem cell pluripotent genes, such as SOX2, OCT4, NANOG, and NESTIN, were dramatically downregulated in a time-dependent manner after RA induced neuritogenesis (Figure 1B), indicating that SH-SY5Y cells can serve a useful cellular model to identify the key regulators of NB differentiation. We transduced SH-SY5Y cells with 573 sgRNA lentiviral supernatants (one sgRNA per well) with puromycin selection on Day 3 to eliminate the uninfected cells (Figure 1C and Supplementary Table S1). Infected cells were cultured for differentiation and imaged at Day 10 with crystal violet staining. After three rounds of screening, we identified PHF20, fibroblast growth factor receptor 2 (FGFR2), myeloid differentiation primary response gene 88 (MYD88), and notch homolog 1 (NOTCH1) involved in the maintenance of the de-differentiated state of SH-SY5Y cells. There are significantly increased neurite density and more differentiated cells in SH-SY5Y cells infected with one of these sgRNAs compared with the control cells (Figure 1D and Supplementary Figure S1). Importantly, FGFR2 and NOTCH1 identified in our screening have been reported to play a role in the maintenance of NB stem cell and malignant phenotypes (Zweidler-McKay, 2008; Katoh and Nakagama, 2014). We further validated the screening results with two sgRNAs against PHF20 using western blotting analysis, and showed marked KO efficiency of PHF20 sgRNA1 and sgRNA2, compared with control sgRNA (Figure 1E). PHF20 KO in SH-SY5Y cells substantially changed cell morphology and downregulated the core pluripotent genes (i.e. SOX2, OCT4, and NANOG), compared with those in control cells (Figure 1F and G). These data suggest that PHF20 is an important regulator to maintain the stemness of NB cells.
Figure 1.
High-throughput screening of key regulators for NB differentiation using a CRISPR/Cas9 sgRNA library. (A) Bright-field microscopy of crystal violet staining of SH-SY5Y cells with and without RA treatment. Neurite outgrowth (arrows) began at Day 3 post-RA treatment. Scale bar, 50 μm. (B) The mRNA expression of SOX2, OCT4, NANOG, and NESTIN of SH-SY5Y cells at 0, 36, and 72 h post-RA treatment. (C) A schematic diagram of the sgRNA library screening system. (D) Heat maps generated from sgRNA library screening of SH-SY5Y cell differentiation analysis. (E) Western blot analysis of PHF20 expression in control cells by non-specific sgRNA and PHF20 KO SH-SY5Y cells by two different PHF20-specific sgRNAs. (F) Crystal violet staining in control cells and PHF20 KO SH-SY5Y cells. Dense neurite networks (arrows) were found in PHF20 KO SH-SY5Y cells. (G) The mRNA expression of SOX2, OCT4, and NANOG in control cells and PHF20 KO SH-SY5Y cells from two different sgRNAs. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
PHF20 is highly expressed, and the expression is associated with the aggressiveness in NB
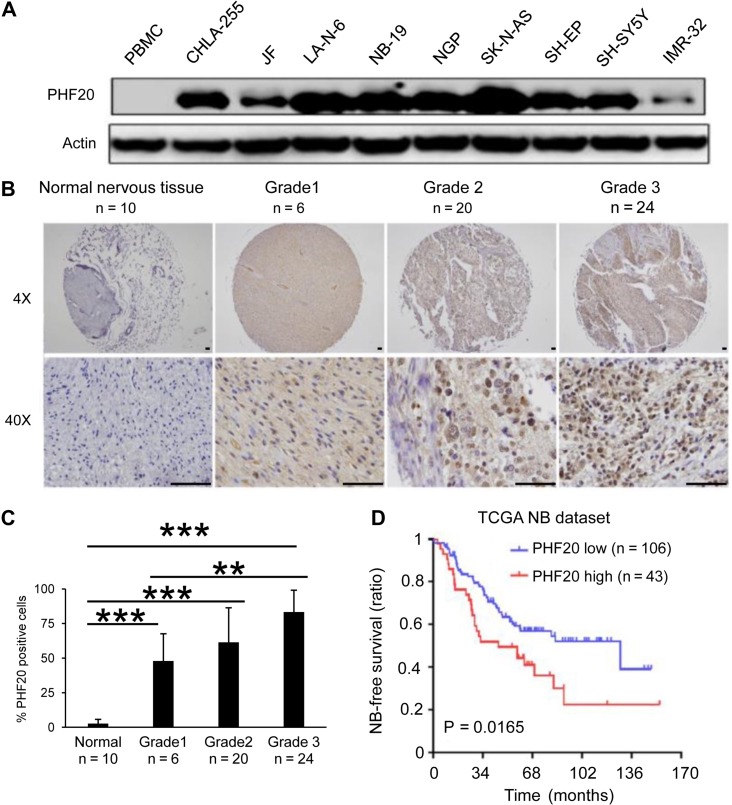
To determine the relationship between PHF20 expression and NB aggressiveness, we investigated the PHF20 expression in the Asgharzadeh NB dataset and found that 92.3% of the 117 NBs had higher PHF20 expression compared with normal tissues (Supplementary Figure S2A). We then determined the protein level of PHF20 in different NB cell lines, including CHLA-255, JF, LA-N-6, NB-19, NGP, SK-N-AS, SH-EP, SH-SY5Y, and IMR-32, and found higher levels of PHF20 expression in all nine NB cell lines, but not in control cells (Figure 2A). Normal peripheral blood mononuclear cells (PBMCs) were used as a control since normal neural cell lines or tissues were not available for western blot analysis. To determine whether PHF20 overexpression is correlated with high histologic grade in NB tissues, we examined PHF20 expression in paraffin-embedded human NB samples by immunohistochemistry, and found a marked increase in the expression of PHF20 in all NB tumor samples when compared with the normal peripheral nerve tissue (Figure 2B) and normal human tissue from different organs (Figure 2C and Supplementary Figure S2B). The association between PHF20 expression in NB and tumor-free survival time of selected patients was analyzed through Kaplan–Meier survival analysis with two different datasets (Figure 2D and Supplementary Figure S2C). In both The Cancer Genome Atlas (TCGA) and Texas Children’s Hospital datasets, high PHF20 expression was inversely correlated with poor median overall survival (OS) of NB patients (P < 0.016) (Figure 2D and Supplementary Figure S2C). Collectively, these results demonstrate the pivotal role of PHF20 in the aggressive behavior of NB and patient overall survival.
Figure 2.
PHF20 is highly expressed in NB and correlates with the poor outcome of NB patients. (A) Western blot analysis of PHF20 expression in nine NB cell lines and normal PBMCs. (B) IHC staining of PHF20 in NB of Grades 1–3 from patients and comparison with normal peripheral nervous tissue. (C) The statistical results showed the proportion of PHF20-positive cells in each group. (D) The association between PHF20 expression in NB and tumor-free survival time of selected patients was analyzed by Kaplan–Meier analysis in TCGA dataset. Scale bar, 50 μm. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
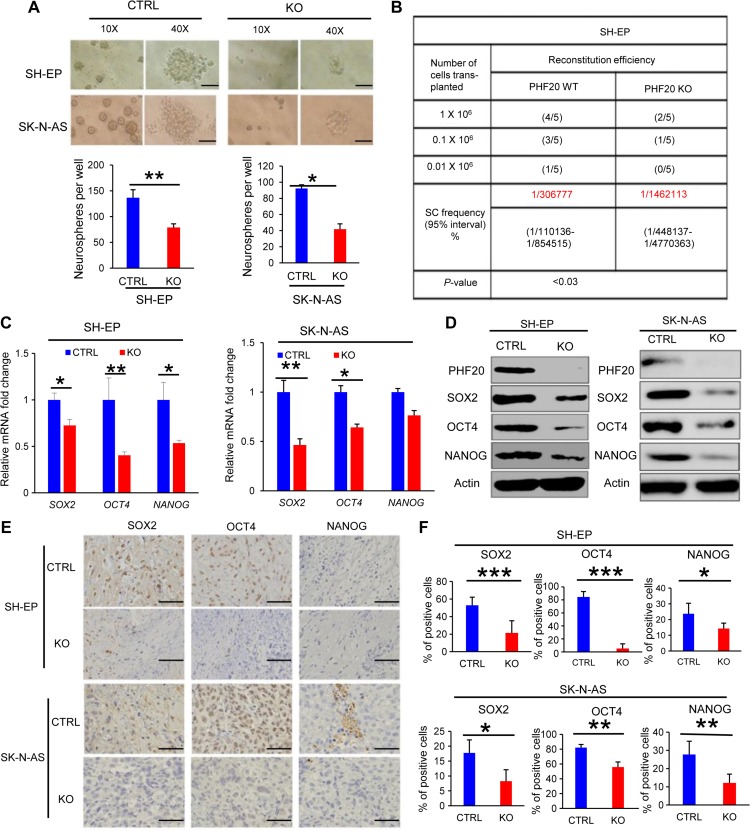
PHF20 increases cellular viability and proliferative capacity of NB cells both in vitro and in vivo
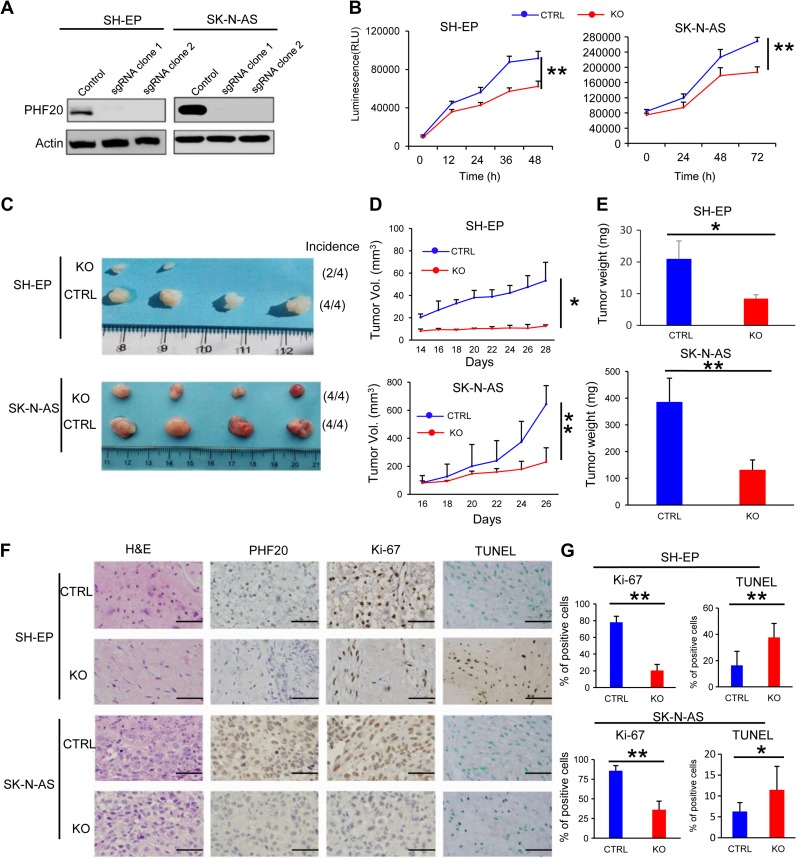
Human NB cell lines can be divided into three categories: N-type (neuroblastic), S-type (substrate-adherent and the non-neuronal), and I-type (the intermediate) (Spengler et al., 1997). N-type (SH-SY5Y) and I-type [BE(2)-C] usually express MYCN at higher levels. Surprisingly, S-type NB cells include both malignant cells such as SK-N-AS and innocent cells such as SH-EP that express low levels of MYCN (Ross et al., 2003), suggesting that other survival mechanisms are important. As N-type and I-type cells express high levels of MYCN, which has been associated to NB aggressiveness. Since our focus was on the role of PHF20, we decided to continue our work using S-type NB cells. To determine the role of PHF20 in S-type NB tumorigenesis, we established PHF20 KO cell clones of SH-EP and SK-N-AS. PHF20 KO in these cells was demonstrated by western blot analysis (Figure 3A) and used for subsequent experiments. Both PHF20 KO SH-EP and SK-N-AS cells showed significantly reduced cell viability compared with cells expressing CRISPR/Cas9 non-specific control sgRNA (Figure 3B). To extend these in vitro observations, we investigated whether PHF20 KO could inhibit the tumorigenic capacity of NB cells in vivo. We injected PHF20 KO SH-EP, PHF20 KO SK-N-AS, and their corresponding control cells subcutaneously into NOD-SCID IL2Rγ-null (NSG) mice. The tumor volumes were measured every other day up to 28 days. PHF20 KO in both SH-EP and SK-N-AS cells remarkably decreased the tumor volume and weight (Figure 3C–E). Furthermore, the xenograft tumors were excised and processed for hematoxylin and eosin (H&E) staining, immunohistochemical staining (IHC), and a terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNEL) assay (Figure 3F). Unlike the high PHF20 expression seen in the control group, IHC staining indicated that PHF20 was deleted in the KO group. The significant reduction in the cell proliferation rate in PHF20 KO cells-derived tumors was further confirmed by measuring Ki-67 levels (Figure 3F and G). The TUNEL assay also showed that the number of apoptotic cells increased in the PHF20 KO groups (Figure 3F and G). Together, these results suggest that PHF20 promotes tumor growth and proliferation, and reduces cell apoptosis.
Figure 3.
PHF20 promotes proliferation of NB cells in vitro and in vivo. (A) Demonstration of ablation of PHF20 in NB PHF20 KO cells by western blotting analysis. PHF20 KO clones were generated with PHF20 sgRNA #1 and #2. (B) A total of 5000 wild-type (WT) and PHF20 KO SH-EP cells and 50000 WT and PHF20 KO SK-N-AS cells were plated in a 96-well plate using 200 μl medium. Cell viability was assayed using CellTiter-Glo®. (C) Representative xenografts excised from NSG mice. The number of mouse xenografts and tumor incidence in each group is noted on the right. (D) Growth of tumors following subcutaneous injection of PHF20 KO or control cells. (E) The tumor weight of subcutaneous xenografts formed by NB WT and PHF20 KO cells is illustrated. (F) Hematoxylin and eosin (H&E) staining and IHC staining of PHF20 and Ki-67, as well as the TUNEL assay of xenografts. (G) The statistical results showing the proportion of Ki-67-positive cells in each field and the proportion of apoptotic cells in the TUNEL assay. Scale bar, 50 μm. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
PHF20 promotes migratory and invasive capacities of NB cells
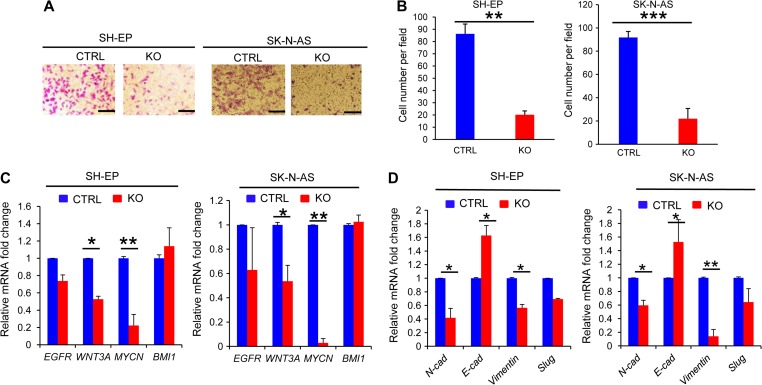
To investigate the effect of PHF20 on cell migration, we first performed the wound healing assay using PHF20 KO SH-EP and PHF20 KO SK-N-AS cells, and found that their migration abilities were significantly reduced compared with control cells (Supplementary Figure S3A). Consistently, we showed that PHF20 KO SH-EP and PHF20 KO SK-N-AS cells had reduced invasive capacity compared with control cells (Figure 4A and B), suggesting that PHF20 is required for the invasive ability of NB cells. Since Wnt/β-catenin (Blanc et al., 2005), MYCN (Kaneko et al., 2015), and epithelial–mesenchymal transition (EMT) signaling (Nieto et al., 2016) drive invasive and metastatic behavior in NB cells, we next examined the expression profiles of these genes on PHF20 KO SH-EP and PHF20 KO SK-N-AS cells. PHF20 KO SH-EP and PHF20 KO SK-N-AS cells remarkably reduced the expression of Wnt3a, Mycn, N-cadherin, and Vimentin genes at the mRNA level, while dramatically increased E-cadherin expression (Figure 4C and D). These findings indicate that PHF20 promotes invasiveness of NB cells by regulating Wnt/β-catenin, MYCN, and EMT signaling expression.
Figure 4.
PHF20 promotes migration and invasion of NB cells. (A) PHF20 KO and its control cells were subjected to transwell matrigel invasion assays. (B) Quantification of migrated cells through Matrigel for each cell line is shown. (C) Expression levels of Egfr, Wnt3a, Mycn, and Bmi1 were analyzed by quantitative real-time PCR (qPCR) in PHF20-deficient and control NB cells. (D) Expression levels of N-cadherin (N-cad), E-cadherin (E-cad), Vimentin, and Slug were analyzed by qPCR in PHF20-deficient and control NB cells. Scale bar, 50 μm. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
PHF20 enhances properties of stem cell-like behavior in NB cells
It has been proposed that tumor-initiating cells (TICs) exhibit stem cell-like properties (Schwitalla et al., 2013). Therefore, we next sought to determine the contribution of PHF20 in the acquisition of cancer stem-like properties. Because neurospheres are the clonal cell clusters of neural stem cells, neurosphere formation assays are well established in neurobiological research (Galli, 2013). Using the neurosphere formation assay, we observed that PHF20 KO markedly decreased clonogenic capacity and sphere size in SH-EP and SK-N-AS cells (Figure 5A). To determine in vivo tumor-initiating efficiency of NB cells, we subcutaneously transplanted PHF20 KO SH-EP cells and control cells into NSG mice at serial dilutions and estimated TIC frequencies using L-Calc software. We found that PHF20 KO cells possessed significantly lower TICs and initiated tumor activity less efficiently compared with the control cells (Figure 5B), suggesting that PHF20 is required for maintaining the TIC-like properties.
Figure 5.
PHF20 confers stem cell-like behavior to NB cells. (A) A tumor sphere formation assay was performed to assess the self-renewal capacity of WT and PHF20 KO cells. Five random wells were photographed. The sphere number was counted after 7 days. (B) A summary of the tumor incidence data for animals after subcutaneous injection of PHF20 KO or control cells is shown. (C) Expression levels of SOX2, OCT4, and NANOG were analyzed by qPCR in PHF20 KO and control NB cells. (D) Western blot analysis of SOX2, OCT4, and NANOG expression in PHF20 KO and control NB cells. β-actin served as a loading control. (E) IHC staining of SOX2, OCT4, and NANOG from xenografts. (F) The statistical results showing proportion of SOX2-, OCT4-, and NANOG-positive cells in each field. Scale bar, 50 μm. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
Pluripotency genes SOX2, OCT4, and NANOG are the most frequently enumerated stem cell factors and are responsible for self-renewal in a variety of tumor types (Cao et al., 2015; Gawlik-Rzemieniewska and Bednarek, 2016; Villodre et al., 2016). The expression of stemness genes, such as SOX2, OCT4, and NANOG, decreased when SH-SY5Y cells were differentiated into neural-like cells induced by PHF20 KO. We reasoned that PHF20 promotes NB stem cell-like properties by regulating the expression of SOX2, OCT4, and NANOG. To test this prediction, we measured gene expression in PHF20 KO SH-EP and PHF20 KO SK-N-AS cells using real-time PCR and western blotting, and found that PHF20 KO dramatically decreased the expression of SOX2, OCT4, and NANOG at the mRNA and protein levels (Figure 5C and D). We further examined the expression of SOX2, OCT4, and NANOG in PHF20 KO SH-EP cell- and control cell-derived tumors. IHC staining results revealed a significant reduction in SOX2, OCT4, and NANOG expression in PHF20 KO SH-EP and PHF20 KO SH-N-AS cell- and control cell-derived tumors (Figure 5E and F), suggesting that PHF20 regulates SOX2, OCT4, and NANOG expression in NB cells. Further, we performed gain-of-function experiments using lentiviral constructs expressing PHF20 in PHF20 KO SH-EP cells, as shown in Supplementary Figure S4A. Importantly, ectopic expression of PHF20 rescued SOX2, OCT4, and NANOG expression in PHF20 KO SH-EP cells (Supplementary Figure S4A), suggesting that PHF20 is necessary to upregulate stem cell core factor expression.
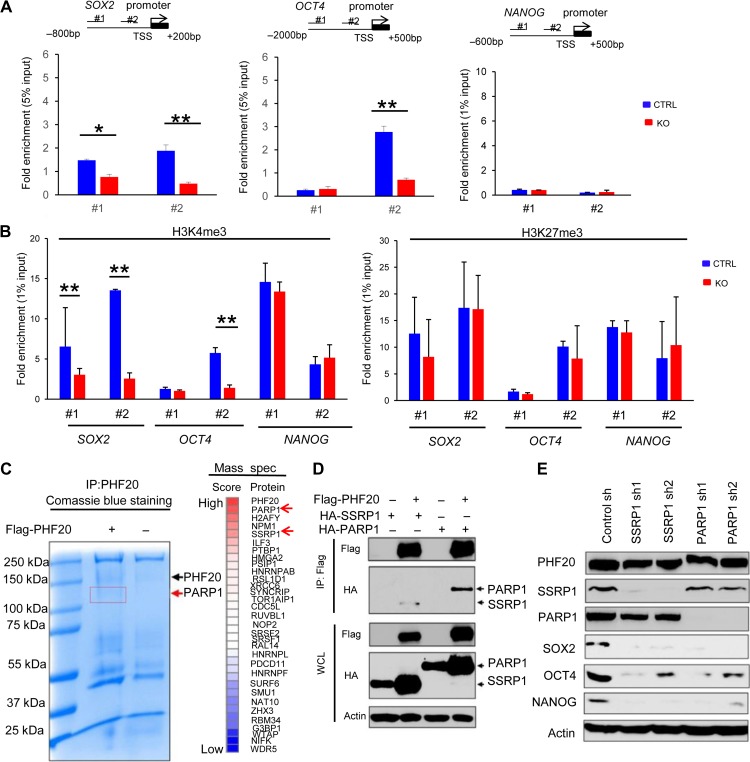
PHF20 regulates the expression of SOX2 and OCT4 through interaction with PARP1 and SSRP1
To further understand how PHF20 regulates SOX2, OCT4, and NANOG expression, we performed chromatin immunoprecipitation–quantitative PCR (ChIP–qPCR) assays using KO SH-EP and control cells. Antibodies against PHF20 were used to pull down the chromatin complex, and two pairs of primers against SOX2, OCT4, and NANOG promoter regions were used. The ChIP–qPCR experiments revealed that PHF20 was strongly bound to SOX2 and OCT4 promoters (Figure 6A). However, PHF20 was not detected at NANOG promoter sites. The trimethylation of H3K4 in promoter regions is associated with active gene expression, while the trimethylation of H3K27 represses gene expression (Barski et al., 2007). We next tested whether the modulation of SOX2, OCT4, and NANOG expression by PHF20 is correlated with H3K4me3 and H3K27me3 modification at their gene promoters. The ChIP–qPCR assay using H3K4me3 antibody showed that decreased SOX2 and OCT4 expression in PHF20 KO SH-EP cells was associated with decreased H3K4me3 levels at the gene promoter regions, compared with the control cells (Figure 6B, left). OCT4 expression was reduced in #2 but not #1 primer set. We postulated that primer #2 was associated with a critical region for PHF20 regulation of OCT4 transcription, while primer #1 did not. Moreover, we did not observe appreciable difference in H3K27me3 occupancy at the gene promoter regions of SOX2 and OCT4 in PHF20 KO cells and control cells (Figure 6B, right), suggesting that the transcriptional activation of SOX2 and OCT4 induced by PHF20 is mainly regulated through modulating H3K4me3 at the gene promoter region in NB cells.
Figure 6.
PHF20 regulates SOX2 and OCT4 expression and H3K4 trimethylation by interacting with PARP1. (A) Analysis of PHF20 binding to the promoter regions of SOX2, OCT4, and NANOG in NB cells by ChIP–qPCR assay with PHF20-specific antibody. The data are presented as fold enrichment relative to input DNA. (B) ChIP–qPCR analysis of H3K4me3 and H3K27me3 of the SOX2, OCT4, and NANOG promoters in PHF20 KO and control cells. (C) Cell lysate of the PHF20 KO cells with ectopic flag-PHF20 expression cell lysate was subjected to immunoprecipitation with PHF20-specific antibody. The resolved proteins were subjected to Coomassie blue staining and excised for mass spectrometry. The top scored proteins that may interact with PHF20 were listed. (D) 293T cells were transfected with Flag-PHF20 or HA-tagged SSRP1 or PARP1. Cell extracts were immunoprecipitated with anti-Flag beads, followed by immunoblotting with anti-HA and anti-Flag antibodies. (E) Western blot analysis of PHF20, SOX2, OCT4, and NANOG expression in SSRP1 and PARP1 knockdown (KD) SH-EP cells using shRNA. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
Because PHF20 is a histone methylation binding protein, we reasoned that PHF20 regulates SOX2 and OCT4 expression by interacting with other epigenetic factors. To identify the candidates of the PHF20 complex in NB cells, we performed PHF20 immunoprecipitation (IP) on cell lysates of PHF20 KO SH-EP cells with PHF20 overexpression. Cell lysates of PHF20 KO SH-EP cells were used as a negative control. Discrete bands stained with Coomassie blue were excised and subjected to mass spectrometric analysis (Figure 6C). More than 200 candidate proteins were identified, with PARP1 yielding the highest score in mass spectrometry data (Figure 6C and Supplementary Table S2). Many of the candidate proteins were nucleic-acid-binding proteins (21%), which is in agreement with the function of PHF20 binding to SOX2 and OCT4 promoters (Supplementary Figure S5A). We subjected the top 30 candidate proteins to the ingenuity pathway analysis (IPA), and found PARP1 to be closely related to the other proteins (Supplementary Figure S5B).
To validate whether PARP1 interacts with PHF20, we immunoprecipitated Flag-tagged PHF20 and detected HA-tagged PARP1 and its partner SSRP1 (Chiou et al., 2013), respectively. Western blot results showed that PHF20 interacts with PARP1 and SSRP1 (Figure 6D). Moreover, we found that PHF20 interacted with WDR5 but not H2AFY (Supplementary Figure S5C). Next, we addressed the question of whether PARP1 indeed had any function in regulating PHF20-driven gene expression. We knocked down PARP1 and SSRP1 using two different shRNAs and analyzed the expression of SOX2, OCT4, and NANOG in SH-EP cells by western blotting, and found that the expression of pluripotency genes significantly decreased in PARP1 and SSRP1 KD SH-EP cells compared with control cells (Figure 6E). Moreover, silencing PARP1 or SSRP1 also inhibited the neurosphere formation capacities of SH-EP cells (Supplementary Figure S5D). ChIP–qPCR experiments revealed that PARP1 directly bound to SOX2 and OCT4 promoters, while SSRP1 only bound to OCT4 promoter (Supplementary Figure S5E). Furthermore, PHF20 binding to SOX2 and OCT4 promoters decreased in PARP1 KD SH-EP cells but not in SSRP1 KD cells, compared with the control cells (Supplementary Figure S5F). These data demonstrate that the binding of PHF20 to SOX2 and OCT4 promoters is mainly dependent on PARP1 in NB cells.
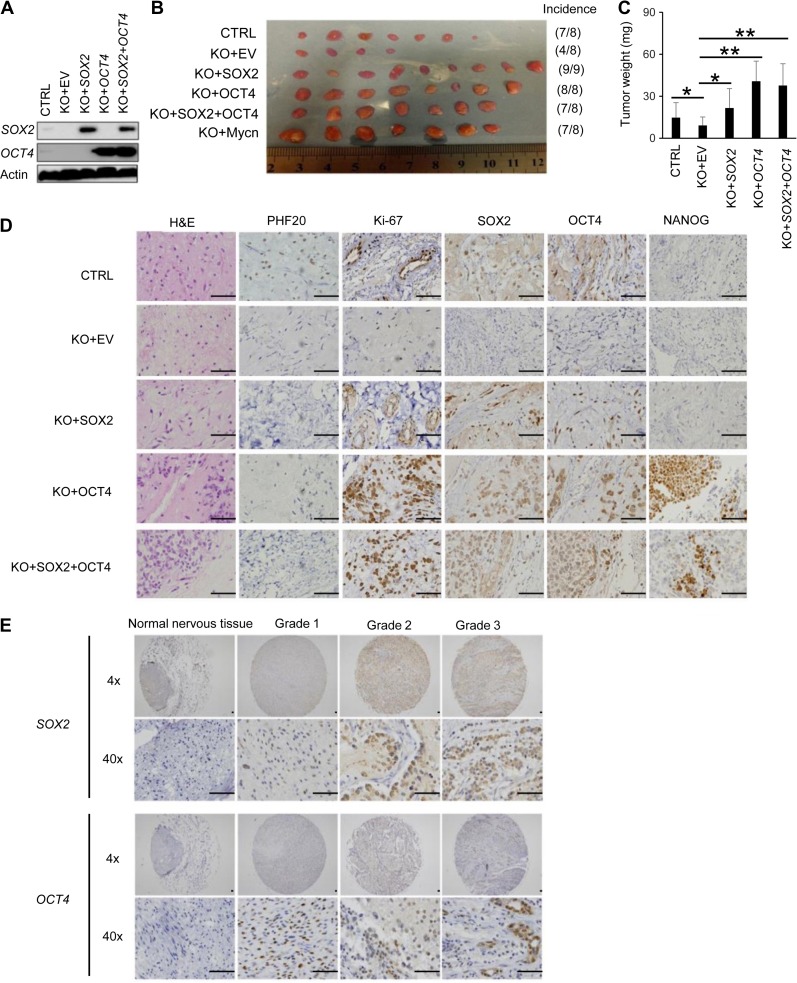
SOX2 and OCT4 play dominant roles in PHF20-induced stemness in NB cells
To test whether the stem cell-like traits conferred by PHF20 were mediated through SOX2 and OCT4, we ectopically expressed SOX2 and OCT4 alone or together in PHF20 KO SH-EP cells. We confirmed the expression of SOX2 and OCT4 in PHF20 KO SH-EP cells using western blot analysis (Figure 7A). Next, we implanted PHF20 KO NB cells, with and without enforced expression of SOX2, OCT4, or both, into NSG mice. SH-EP cells with empty vector and PHF20 KO SH-EP cells with MYCN overexpression were used as negative and positive controls, respectively. We observed that overexpressing SOX2 and OCT4, alone or together, significantly rescued the tumor incidence in the xenograft model induced by PHF20 KO SH-EP cells (Figure 7B). Further, PHF20 KO cells overexpressing SOX2 and OCT4 generated significantly larger tumors compared to PHF20 KO cells alone (Figure 7C). Using IHC staining, we showed that SOX2 and OCT4 were indeed highly expressed in a variety of groups (Figure 7D). Moreover, SOX2 and OCT4 overexpression increased the percentage of Ki-67-positive cells compared to the PHF20 KO group (Figure 7D). These results clearly demonstrate that SOX2 and OCT4 are critical mediators of PHF20-promoted stem cell-like phenotypes in NB cells.
Figure 7.
OCT4 and SOX2 play dominant roles in PHF20-induced stemness in NB cells. (A) Western blot analysis of SOX2 and OCT4 overexpressed either individually or in combination in PHF20 KO cells. (B) Representative xenografts excised from different groups of NSG mice are shown. Incidence was calculated by the number of tumors formed divided by total number of mice for each group. (C) The average tumor weight of varied groups. (D) H&E and IHC staining of PHF20, Ki-67, SOX2, OCT4, and NANOG expression in different groups. (E) IHC staining of SOX2 and OCT4 in NB tissue samples of Grades 1–3 from patients and in normal peripheral nervous tissue. Scale bar, 50 μm. Data are plotted as mean ± SD of three independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 compared with controls using Student’s t-test.
To further substantiate these findings, we next analyzed SOX2 and OCT4 expression in human NB tissue arrays, and found both SOX2 and OCT4 overexpression in patients with NB tumor grade ranging from 1 to 3 (Figure 7E). Therefore, our study identifies PHF20 as a key regulator of NB growth and metastasis through upregulation of SOX2 and OCT4 expression, thus serving as an important biomarker for NB diagnosis and therapeutics.
Discussion
The current therapy of high-risk NB patients includes surgery, intensive myeloablative chemotherapy with autologous peripheral blood stem cell reinfusion, retinoid treatment, and antibody therapy (Cheung and Dyer, 2013). Even with intensive therapies, due to the aggressive nature of this disease, most NB patients relapse with therapy-resistant tumors (Meacham and Morrison, 2013). Incomplete understanding of NB differentiation has hampered the development of new therapeutic approaches for aggressive NB. One potential therapeutic option is to develop immunotherapy against NB by targeting overexpressed proteins like PHF20 (Fischer et al., 2001). Our present study has identified a novel function of PHF20 in promoting an essential characteristic of malignant NB, namely the cancer stemness. To elucidate the mechanisms of how PHF20 promotes stem cell-like traits in NB cells, we show that SOX2 and OCT4 are regulated by PHF20 and function as effective mediators of PHF20-induced NB initiation and progression.
Our recent study shows that PHF20 functions as a key regulator of stem cell self-renewal and cellular reprogramming (Zhao et al., 2013). The role and molecular mechanisms by which PHF20 contributes to tumor stem cell-like properties remain largely unknown. Originally, discovered as an autoantibody in glioblastoma patients (Pallasch et al., 2005), PHF20 has been found to be abundantly expressed in several other cancer types (Pallasch et al., 2005; Bankovic et al., 2010; Zaatar et al., 2012). Furthermore, PHF20 plays a vital role in carcinogenesis by significantly enhancing the self-renewal and tumor-initiating capability of lung cancer cells (Klein et al., 2016). In this study, we demonstrated that the elevated PHF20 expression promotes tumor cell growth and proliferation and is inversely correlated with poor outcome in NB patients.
It has been shown that PHF20 exhibits high selectivity for H3K4me2 as an epigenetic reader for further modification (Klein et al., 2016). Moreover, PHF20 could recruit the H3K4 methyltransferase complex to modulate H3K4me3 of the OCT4 promoter during cellular reprogramming through interaction of WDR5 (Zhao et al., 2013). Our current study showed that deletion of PHF20 expression significantly decreased the trimethylation status of H3K4 at the SOX2 and OCT4 gene promoters. This process consequently reduced expression of NANOG, as OCT4, SOX2, and NANOG (OSN) form a positive feedback regulation loop at the transcriptional level. Our data identified a previously unknown epigenetic connection between PHF20 and stem cell core factors (i.e. OSN) in NB. Furthermore, PHF20 interacts with PARP1 to promote the accumulation of H3K4me3 for epigenetic control of SOX2 and OCT4 activity in NB cells. PARP1 has been reported to control H3K4me3 through PARylation to exclude histone demethylase KDM5B (Krishnakumar and Kraus, 2010). Previous studies support a role of PARP1 in the regulation of pluripotency networks in stem cells (Roper et al., 2014). In the absence of PARP1, embryonic stem (ES) cells exhibit a decrease in ground state pluripotency (Doege et al., 2012). PARP1 can be recruited to the NANOG and Esrrb loci to establish early epigenetic marks during somatic cell reprogramming (Doege et al., 2012). In addition, the epigenetic modulation function of PARP1 is involved in regulating expression for a wide variety of genes (Jiang et al., 2015). As such, PARP1 may be a prerequisite for the NB stem-cell fate. Further studies are needed to investigate the role of PARP1 in NB. Given the low frequency of mutations in NB, PHF20-induced epigenetic regulation through its interaction with PARP1 may play an essential role in the control of NB stemness.
We show that expression of SOX2 and OCT4 is correlated to the clinical stages of NB. SOX2 is an important regulator of cellular processes for cancer development, including Wnt/β-Catenin and EMT signaling (Li et al., 2013). PHF20 ablation reduced SOX2 expression and blocked the Wnt and EMT-induced expression of N-cadherin and Vimentin. We propose that the PHF20−SOX2 axis is involved in activation of the Wnt signaling and the metastatic EMT factors for the maintenance of NB invasive phenotype. These findings provide a mechanistic framework explaining clinical observations that NB tumors expressing high levels of PHF20 also highly express SOX2 and OCT4 and are associated with poor patient survival. Previous studies have shown that OCT4 induced MYCN expression in human NB cells (Kaneko et al., 2015). In this study, we found that PHF20 ablation decreased transcription of both OCT4 and MYCN. Thus, OCT4 may serve as a critical link between PHF20 and MYCN in NB. It has been established that MYCN-positive NB cells are potently resistant to differentiation (Loven et al., 2010). Therefore, functional interplay between PHF20 and MYCN may contribute to maintaining the stem cell-like status of NB cells.
Based our findings, we propose a working model showing that PHF20, together with PARP1, modulates the trimethylation of H3K4 of SOX2 and OCT4 promoters, leading to their activation and subsequent induction of NANOG. This further alters the expression of Wnt, MYCN, and EMT signaling (Supplementary Figure S6). Collectively, these events direct NB cell malignancy as evidenced by outgrowth, increased cell motility and invasion, and upregulated capacity to self-renew. Our findings provide novel therapeutic targets by pharmacological inhibition of PHF20 or for developing immunotherapy through targeting PHF20 against NB.
Materials and methods
Cell culture and reagents
Human NB cell lines were grown in Gibco® RPMI 1640 medium containing 10% fetal bovine serum (FBS) at 37°C in a humidified 5% CO2 atmosphere.
CRISPR/Cas9 sgRNA library screening
SH-SY5Y cells were cultured in 96-well plates at 1 × 103 cells per well. After 24 h, the cells were infected with lentivirus harboring different sgRNAs at appropriate concentrations. For each differentiation screening, three wells were grouped together as one triplicate. Cells were grown in RPMI 1640 medium containing 2 μg/ml puromycin for 3 days, then switched to fresh RPMI 1640 medium. After growing for an additional 7 days, the puromycin resistant cells were stained with crystal violet to ascertain cell morphology and degree of differentiation. Differentiated cells, those with a neurite length of greater than 10 μm, were counted using the Metamorph™ software.
Generation of PHF20 KO cell lines
SH-EP or SH-N-AS cells were stably transduced with a PHF20 sgRNA and cloned by limiting dilution cloning. Briefly, cells were plated at a density of 2 × 105 cells per 6-well plate. Cells were individually transduced with non-specific sgRNA or PHF20 sgRNA expression lentivirus. Two days after transduction, cells were cultured with the 2 μg/ml puromycin for 3 days. Then, the cells were re-seeded at a density of 0.3 cells per well in 96-well plates and grown in medium containing 2 μg/ml puromycin. After 3 weeks, 10−30 monoclones per sgRNA were picked and expanded. The PHF20 KO efficiency of monoclones was evaluated by western blot analysis.
PARP1 and SSRP1 shRNA gene silencing
PARP1, SSRP1, and non-specific control lentiviral shRNAs were purchased from GIPZ shRNA library (GE Dharmacon). PARP1 shRNA 1 clone ID: V2LHS_196007; PARP1 shRNA 2 clone ID: V2LHS_201984; SSRP1 shRNA1 clone ID: V2LHS_153561; SSRP1 shRNA2 clone ID: V3LHS_363710. SH-EP cells were transduced with lentivirus harboring different shRNAs. Prior to use, shRNA-positive cells were selected for by culturing in medium containing 2 μg/ml puromycin for 1 week.
Cell viability assay
CellTiter-Glo® Luminescent Cell Viability Assay (Promega) was used to determine NB cell viability, as per manufacturer’s directions. Cells were seeded in 96-well plates at a density of 5 × 103 SH-EP cells/well or 5 × 104 SK-N-AS cells/well, and then were incubated at 37°C in a humidified 5% CO2 atmosphere. The culture medium was discarded at 0, 12, 24, 36, and 48 h; cell lysis was induced by adding 40 μl of CTG solution to each well and incubating for 2 min at 37°C on an orbital shaker. Staining intensity in the medium was determined by measuring absorbance (optical density, OD) at 450 nm.
Wound healing assay
Cells were seeded in 6-well plates and grown to 90% confluency. The cell monolayers were scraped using a sterile plastic tip, followed by culturing in RPMI 1640 medium with 1% FBS at 37°C in a humidified 5% CO2 atmosphere. Micrographs were taken to assess cell migration at 0, 12, 24, 36, and 48 h.
Transwell invasion and migration assay
The invasiveness of NB cells was assessed by their ability to pass through Corning Matrigel Matrix (CORNING)-coated Transwell inserts (Millipore). The upper surface of the polycarbonic membranes (8.0 μm pore size) of the transwell chambers was coated with Matrigel (1:4 diluted with RPMI 1640). Cells (1 × 104) in 100 μl of serum-free RPMI 1640 medium were seeded into the upper compartments of the chambers. The lower compartments of the chambers were filled with 500 μl of RPMI 1640 containing 10% FBS. After 48 h, invasive cells that migrated from the Matrigel to the lower surface of the filters were fixed in 70% ethanol, stained with 0.2% crystal violet, and counted under an inverted microscope at 100× magnification. Cell invasion was determined by averaging the number of cells counted in four randomly selected visual fields per filter.
Neurosphere assay
Cells were seeded at a concentration of 2000 cells/100 μl in complete neural stem cell (NSC) basal medium [9:1 mixture of NSC basal medium and NSC proliferation supplement containing 20 ng/ml EGF, 10 ng/ml basic fibroblast growth factor (bFGF), and 1 μl/ml of 0.2% heparin] in each well of a 24-well ultralow-attachment plate (Corning Life Sciences). One milliliter of the NSC basal medium was used in each of the 24 wells. After 7 days of incubation, sphere number and sphere size were counted and analyzed.
Real-time PCR and ChIP−qPCR analysis
Real-time PCR and ChIP assay were performed with specific kit. See Supplementary material for details.
Western blot, IP, and mass spectrometry
The cells were lysed in RIPA buffer, and analyzed. See Supplementary material for details.
Ingenuity pathway analysis
The top 30 candidate genes identified by mass spectrometry were uploaded into the IPA software (Qiagen). The core analysis function included in the software was used to interpret the differentially expressed data, including gene networks. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathway Knowledge Base (IPKB). Nodes (proteins) and edges (the biological relations between the nodes) were generated on the basis of their functional and biological connectivity. The length of an edge reflects the evidence in the literature supporting that node-to-node relation.
Human datasets and survival analysis
Three publically accessible online data repositories for cancer were used in this study: Gene Expression Omnibus (GEO), The Cancer Genome Atlas (TCGA), and Texas Children’s Hospital. For the determination of PHF20 expression in human NB tissue and normal adrenal tissue, we analyzed series GSE3446, titled ‘Gene expression profiles of primary tumors from patients with metastatic NB lacking MYCN amplification’ (n = 117). For the Kaplan–Meier analysis, we used the Texas Children’s Hospital dataset (Houston, TX; n = 88) and the TCGA dataset (RNA-seq data offered by Therapeutically Applicable Research to Generate Effective Treatments [TARGETS], n = 149). The TCGA dataset can be accessed via http://cancergenome.nih.gov/. Briefly, the samples within each dataset were sorted according to PHF20 expression. Based on the X-tile cutoff expression value, we divided the samples into two groups, PHF20 low and PHF20 high. All cutoff expression levels and their resulting groups were analyzed for survival. We selected the strongest significance based on the P-value and the corresponding cutoff value to generate the Kaplan–Meir graphs.
Subcutaneous tumor model
Six-week-old male and female NSG mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in individually ventilated microisolator cages. All animal experiments were approved by the Institutional Animal Care and Use Committee (Houston Methodist Research Institute). See Supplementary material for details.
Immunohistochemistry
Paraffin-embedded human NB and peripheral nerve tissue array (MC602) was purchased from US Biomax. The study cohort consisted of 30 cases (60 cores), comprised of 5 cases with normal peripheral nerve tissue and 25 cases with tumor lymph node metastasis (TNM) grading that ranged from 1 to 3. The xenograft tissues were formalin-fixed, processed, and paraffin-embedded. The H&E staining was performed in the Cancer Pathology Laboratory at Houston Methodist Hospital. Antigens were retrieved by autoclaving in 0.01 mol/L sodium citrate buffer (pH 6.0) at 121°C, 20 psi for 3−5 min. Endogenous peroxidase activity and non-specific binding sites were blocked using 3% hydrogen peroxide and 10% goat serum, respectively. The blocked sections were incubated overnight at 4°C with primary antibody in 1% BSA in phosphate buffer saline with Tween 20 (PBST), followed by 1 h incubation with secondary antibody. The slides were stained with diaminobenzidine (DAB) for 2 min, counterstained with hematoxylin, and mounted with Immuno-mount (Thermo Fisher Scientific). The scoring criterion was the average percentage of positively stained cells counted in 10 randomly selected visual fields.
TUNEL assay
TUNEL assay was performed using TumorTACS™ In Situ Apoptosis Detection Kit 4815-30-K (TREVIGEN) as per manufacturer’s instructions. Briefly, 4-μM thick formalin-fixed, paraffin-embedded tissue sections were de-paraffinized and re-hydrated. Endogenous peroxidase activity was quenched by hydrogen peroxide, and tissue protein was hydrolyzed with proteinase K. All sections were incubated with 50 μl/sample labeling reaction mix at 37°C for 1 h in a humidity chamber. After labeling reaction stopped, samples were covered with Strep-HRP solution and incubated for 10 min at 37°C, treated with DAB solution, and counterstained with methyl green. The scoring criterion was the average percentage of positively stained cells counted in ten randomly selected visual fields.
Statistical analysis
All statistical analyses were performed using GraphPad Prism version 5.0 (GraphPad Software). Data are presented as mean ± SD of three independent experiments. Association among the expression levels of PHF20 and SOX2, OCT4 signals in human NB tissues and tumor stages was analyzed using Spearman rank correlation coefficient test. Comparisons between two groups were performed using two-sided Student’s t-test. For all tests, a P-value <0.05 was considered statistically significant.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Supplementary Material
Acknowledgements
We would like to thank Drs David S. Baskin and Martyn A. Sharpe in Houston Methodist Hospital for their critical support. We are grateful to Dr Jianhua Yang at Baylor College of Medicine for kindly providing all NB cell lines as a gift.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81572766 and 31771630), the National Key Research and Development Program of China (2017YFA0103800), Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S029), Guangdong Natural Science Foundation (2016A030313215 and 2016A030313238), SYSU Young Teachers Training Program (16YKZD14) and grants (CA101795 and 1U54CA210181) from U.S. National Cancer Institute, National Institutes of Health (NIH), DOD (W81XWH-16-1-0417), and CPRIT (DP150099, RP170537, and RP150611).
Conflict of interest
none declared.
Author contributions
W.L. and W.Z. performed most experiments and data analysis. B.N., J.H., and J.C. conducted bioinformatic analysis of mass spectrometry data. L.L. and Q.M. prepared the plasmids and lentiviral vectors. C.X. and H.Y.W. conducted the animal experiments. Q.L. provided suggestions and experimental designs. W.L., W.Z., and R.-F.W. wrote the manuscript. R.-F.W. provided the experimental designs and supervision of the entire project.
References
- Bankovic J., Stojsic J., Jovanovic D., et al. (2010). Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer 67, 151–159. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., et al. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837. [DOI] [PubMed] [Google Scholar]
- Blanc E., Goldschneider D., Douc-Rasy S., et al. (2005). Wnt-5a gene expression in malignant human neuroblasts. Cancer Lett. 228, 117–123. [DOI] [PubMed] [Google Scholar]
- Cao S.G., Ming Z.J., Zhang Y.P., et al. (2015). Sex-determining region of Y chromosome-related high-mobility-group box 2 in malignant tumors: current opinions and anticancer therapy. Chin. Med. J. (Engl.) 128, 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung N.K., and Dyer M.A. (2013). Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat. Rev. Cancer 13, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou S.H., Jiang B.H., Yu Y.L., et al. (2013). Poly(ADP-ribose) polymerase 1 regulates nuclear reprogramming and promotes iPSC generation without c-Myc. J. Exp. Med. 210, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmielecki J., Bailey M., He J., et al. (2017). Genomic profiling of a large set of diverse pediatric cancers identifies known and novel mutations across tumor spectra. Cancer Res. 77, 509–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doege C.A., Inoue K., Yamashita T., et al. (2012). Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 488, 652–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Struss A.K., Hemmer D., et al. (2001). Glioma-expressed antigen 2 (GLEA2): a novel protein that can elicit immune responses in glioblastoma patients and some controls. Clin. Exp. Immunol. 126, 206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli R. (2013). The neurosphere assay applied to neural stem cells and cancer stem cells. Methods Mol. Biol. 986, 267–277. [DOI] [PubMed] [Google Scholar]
- Gawlik-Rzemieniewska N., and Bednarek I. (2016). The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol. Ther. 17, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., and Weiss W.A. (2013). Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 3, a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoueix-Lerosey I., Schleiermacher G., Michels E., et al. (2009). Overall genomic pattern is a predictor of outcome in neuroblastoma. J. Clin. Oncol. 27, 1026–1033. [DOI] [PubMed] [Google Scholar]
- Jiang B.H., Chen W.Y., Li H.Y., et al. (2015). CHD1L regulated PARP1-driven pluripotency and chromatin remodeling during the early-stage cell reprogramming. Stem Cells 33, 2961–2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Suenaga Y., Islam S.M., et al. (2015). Functional interplay between MYCN, NCYM, and OCT4 promotes aggressiveness of human neuroblastomas. Cancer Sci. 106, 840–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M., and Nakagama H. (2014). FGF receptors: cancer biology and therapeutics. Med. Res. Rev. 34, 280–300. [DOI] [PubMed] [Google Scholar]
- Klein B.J., Wang X., Cui G., et al. (2016). PHF20 readers link methylation of histone H3K4 and p53 with H4K16 acetylation. Cell Rep. 17, 1158–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R., and Kraus W.L. (2010). PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol. Cell 39, 736–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xu Y., Chen Y., et al. (2013). SOX2 promotes tumor metastasis by stimulating epithelial-to-mesenchymal transition via regulation of WNT/β-catenin signal network. Cancer Lett. 336, 379–389. [DOI] [PubMed] [Google Scholar]
- Louis C.U., and Shohet J.M. (2015). Neuroblastoma: molecular pathogenesis and therapy. Annu. Rev. Med. 66, 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loven J., Zinin N., Wahlstrom T., et al. (2010). MYCN-regulated microRNAs repress estrogen receptor-alpha (ESR1) expression and neuronal differentiation in human neuroblastoma. Proc. Natl Acad. Sci. USA 107, 1553–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham C.E., and Morrison S.J. (2013). Tumour heterogeneity and cancer cell plasticity. Nature 501, 328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar J.J., Domingo-Fernandez R., Ebus M.E., et al. (2012). LIN28B induces neuroblastoma and enhances MYCN levels via let-7 suppression. Nat. Genet. 44, 1199–1206. [DOI] [PubMed] [Google Scholar]
- Mu P., Zhang Z., Benelli M., et al. (2017). SOX2 promotes lineage plasticity and antiandrogen resistance in TP53- and RB1-deficient prostate cancer. Science 355, 84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., et al. (2016). EMT: 2016. Cell 166, 21–45. [DOI] [PubMed] [Google Scholar]
- Olsson M., Beck S., Kogner P., et al. (2016). Genome-wide methylation profiling identifies novel methylated genes in neuroblastoma tumors. Epigenetics 11, 74–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallasch C.P., Struss A.K., Munnia A., et al. (2005). Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. Int. J. Cancer 117, 456–459. [DOI] [PubMed] [Google Scholar]
- Powers J.T., Tsanov K.M., Pearson D.S., et al. (2016). Multiple mechanisms disrupt the let-7 microRNA family in neuroblastoma. Nature 535, 246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper S.J., Chrysanthou S., Senner C.E., et al. (2014). ADP-ribosyltransferases Parp1 and Parp7 safeguard pluripotency of ES cells. Nucleic Acids Res. 42, 8914–8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R.A., Biedler J.L., and Spengler B.A. (2003). A role for distinct cell types in determining malignancy in human neuroblastoma cell lines and tumors. Cancer Lett. 197, 35–39. [DOI] [PubMed] [Google Scholar]
- Schwitalla S., Fingerle A.A., Cammareri P., et al. (2013). Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152, 25–38. [DOI] [PubMed] [Google Scholar]
- Singovski G., Bernal C., Kuciak M., et al. (2016). In vivo epigenetic reprogramming of primary human colon cancer cells enhances metastases. J. Mol. Cell Biol. 8, 157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.N., Singh A.M., and Dalton S. (2010). Myc represses primitive endoderm differentiation in pluripotent stem cells. Cell Stem Cell 7, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spengler B.A., Lazarova D.L., Ross R.A., et al. (1997). Cell lineage and differentiation state are primary determinants of MYCN gene expression and malignant potential in human neuroblastoma cells. Oncol. Res. 9, 467–476. [PubMed] [Google Scholar]
- Suva M.L., Riggi N., and Bernstein B.E. (2013). Epigenetic reprogramming in cancer. Science 339, 1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Sipp D., and Enomoto H. (2013). Tissue interactions in neural crest cell development and disease. Science 341, 860–863. [DOI] [PubMed] [Google Scholar]
- Villodre E.S., Kipper F.C., Pereira M.B., et al. (2016). Roles of OCT4 in tumorigenesis, cancer therapy resistance and prognosis. Cancer Treat. Rev. 51, 1–9. [DOI] [PubMed] [Google Scholar]
- Zaatar A.M., Lim C.R., Bong C.W., et al. (2012). Whole blood transcriptome correlates with treatment response in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 31, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Li Q., Ayers S., et al. (2013). Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 152, 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweidler-McKay P.A. (2008). Notch signaling in pediatric malignancies. Curr. Oncol. Rep. 10, 459–468. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.