Extended Data Figure 3. Functional characterization of SMAD2/3 transcriptional and epigenetic cofactors during endoderm differentiation.
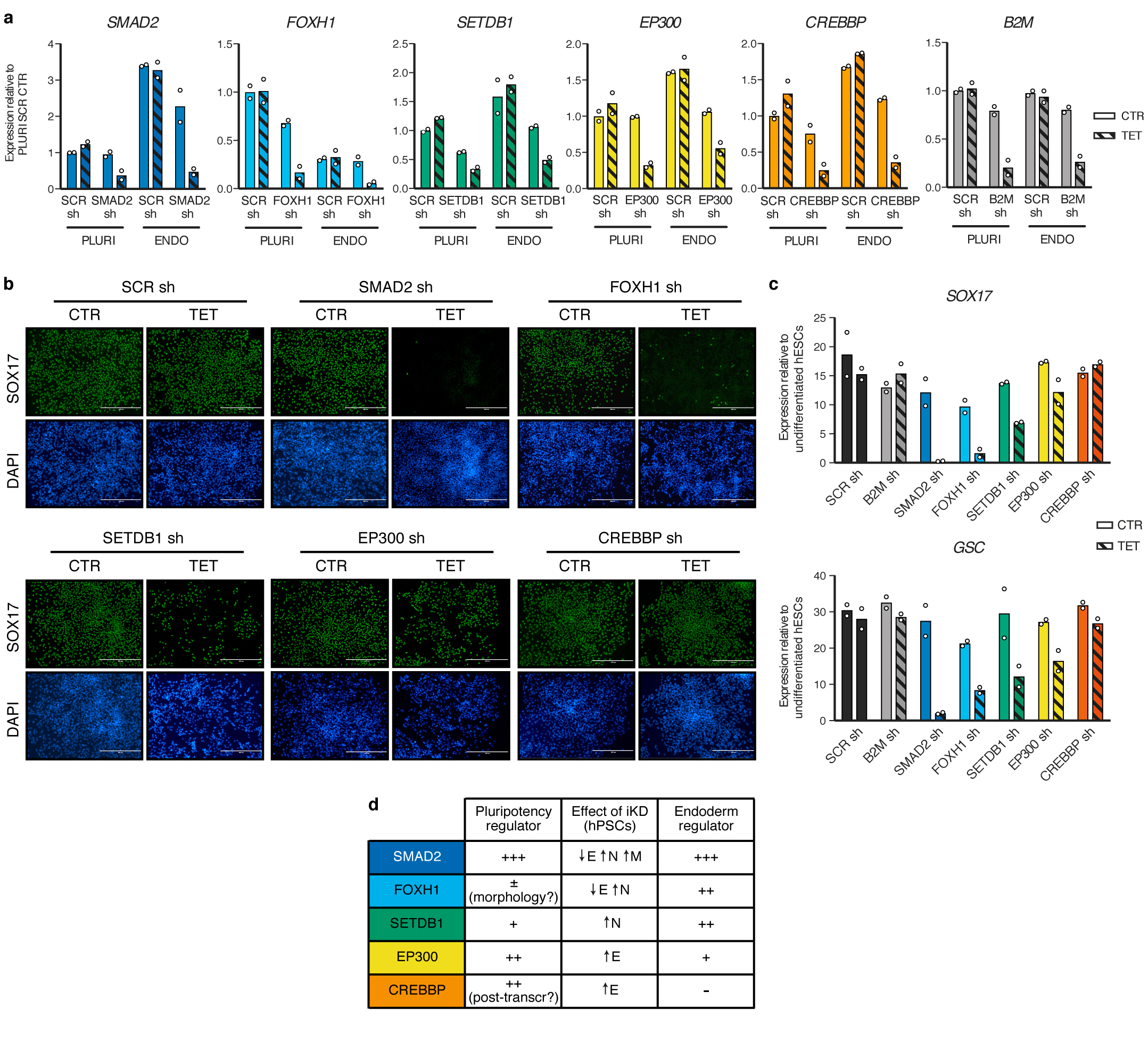
(a) qPCR validation of inducible knockdown (iKD) hESCs in pluripotency (PLURI) and following endoderm differentiation (ENDO). Pluripotent cells were cultured in absence (CTR) or presence of tetracycline (TET) for 6 days. For endoderm differentiation, tetracycline treatment was initiated in undifferentiated hESCs for 3 days in order to ensure gene knockdown at the start of endoderm specification, and was then maintained during differentiation (3 days). For each gene, the shRNA resulting in the strongest level of knockdown in hPSCs was selected (refer to Extended Data Fig. 2). Expression is shown as normalized to the average level in pluripotent hESCs carrying a scrambled (SCR) control shRNAs and cultured in absence of tetracycline. The mean is indicated, n=2 independent clonal pools. (b) Immunofluorescence for the endoderm marker SOX17 following endoderm differentiation of iKD hESCs expressing the indicated shRNAs (sh) and cultured as described in panel a. DAPI shows nuclear staining. Scale bars: 400μm. Results are representative of two independent experiments. (c) qPCR following endoderm differentiation of iKD hESCs. The mean is indicated, n=2 independent clonal pools. (d) Table summarizing the phenotypic results presented in Extended Data Fig. 2 and in this figure. E: endoderm; N: neuroectoderm; M: mesoderm.
