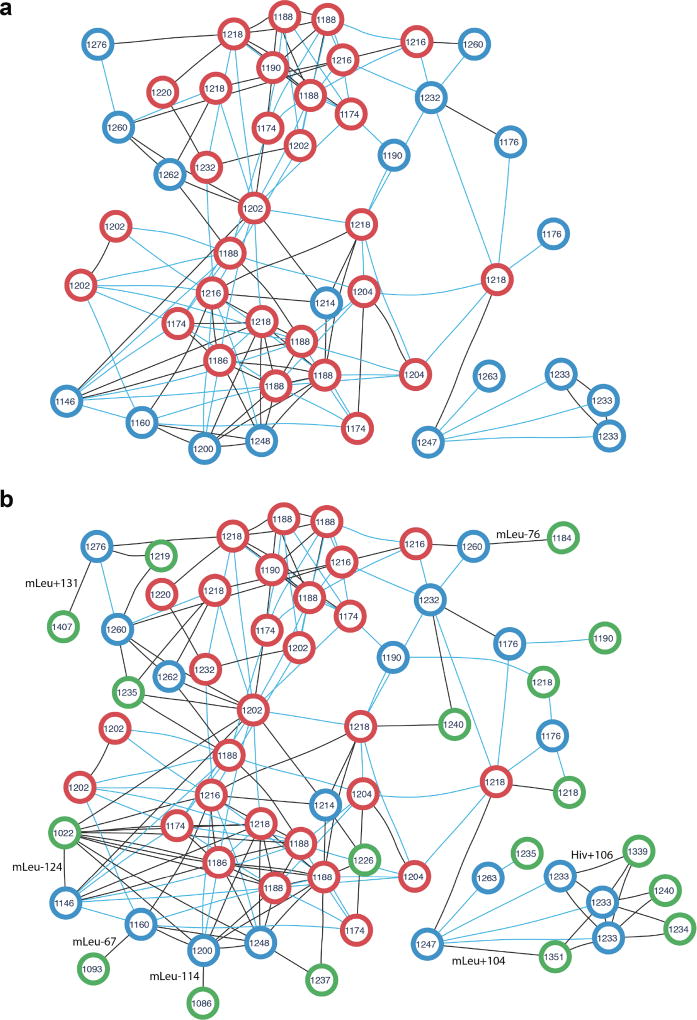
Figure 3. Peptide network of the cyclosporin family in the PNPdatabase (a) extended by the newly identified cyclosporin variants (b).
The peptide network was constructed for 47 known cyclosporins using the GNPS interface (the Molecular Networking workflow2). Each node represents theoretical spectrum of a cyclosporin variant, the number inside each node stands for monoisotopic mass in Da rounded to integers. Two nodes are connected by an edge if the corresponding theoretical spectra are similar (cosine score at least 0.8). Blue edges corresponds to characteristic mass shifts of 14, 16, 28, 32, and 42 Da, the remaining edges are black. (a) Peptide network of cyclosporin variants present in the PNPdatabase. Red nodes are 29 cyclosporins identified in SpectraGNPS as known PNPs (both by DEREPLICATOR and VarQuest). Blue nodes are theoretical spectra of the rest 18 PNPs which are not present in GNPS in their known form and added to the network for the sake of completeness. (b) Peptide network of known and novel cyclosporin variants. Green nodes are theoretical spectra of 18 novel variants identified by VarQuest in GNPS. Each novel variant is the most statistically significant identification of the corresponding blue node (an absent known cyclosporin PNP). Likely insertions/deletions are shown on corresponding edges, Hiv stands for Hydroxyisovaleric acid, mLeu stands for methylated leucine.