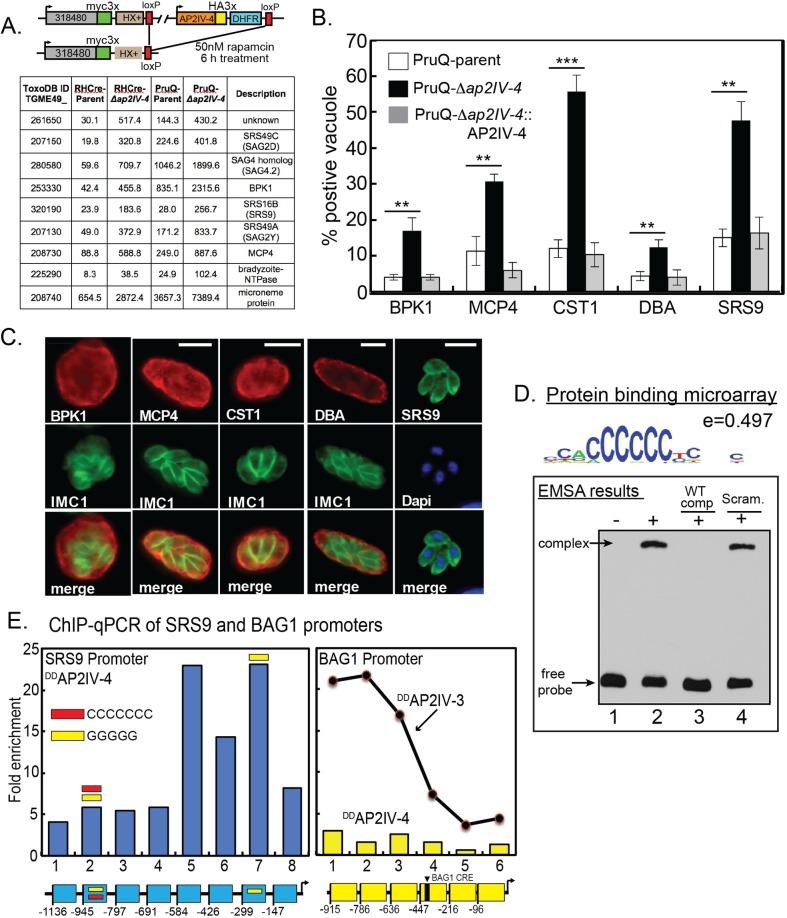
Fig 3. Tissue cyst wall and bradyzoite surface proteins accumulate in tachyzoites lacking AP2IV-4.
(A.) Schematic representation of the AP2IV-4 knockout strategy in the RHCre strain. Insertion of loxP sites surrounding the AP2IV-4 locus was accomplished by sequential epitope tagging of TGGT1_318480 and AP2IV-4 genes utilizing the indicated tags and selectable markers. A similar strategy was also used to delete the AP2IV-4 gene in the PruQ-parent strain (S3B Fig). Active Cre-recombinase excises the floxed AP2IV-4 gene in the dual-tagged RHCre and PruQ-strains, which are the parents referred to in this study. Microarray analysis: selected mRNAs altered by the knockout of the AP2IV-4 gene in RHCre- and PruQ-Δap2IV-4 parasites; average RMA values for RHCre- or PruQ-parent (carrying AP2IV-4floxed) versus Δap2IV-4 knockout transgenic strains are shown (complete results in S1 Dataset). Note the higher baseline expression of mRNAs in the PruQ- versus the RHCre-parent strains. (B. and C.) Average fraction of vacuoles grown under tachyzoite conditions showing DBA+ tissue cyst walls and expression of bradyzoite cyst wall proteins BPK1, MPC4, CST1 and the bradyzoite-specific surface antigen SRS9 in the PruQ-parent, PruQ-Δap2IV-4 knockout, and PruQ-Δap2IV-4::AP2IV-4 complemented strains with representative IFA images in (C.) Statistical significance of pairwise parent versus knockout results is indicated (**, p<0.01; ***, p<0.001). Positive staining for each antigen and DBA+ vacuoles were quantified in triplicate by counting 100 vacuoles from randomly selected microscopic fields. Note also the normal localization of BPK1, CST1, and MCP4 proteins in cyst walls also stained by DBA and the parasite surface localization of SRS9. Unlike bradyzoite heterogenous expression patterns, SRS9 in PruQ-Δap2IV-4 parasites always showed uniform intravacuolar expression consistent with the synchronous replication of tachyzoites. Scale bar = 5 μm. IFA analysis and quantification of these same bradyzoite proteins in RHCre-Δap2IV-4 parasites is presented in supplemental (S4 Fig). (D.) To determine whether the single AP2 domain in AP2IV-4 is capable of binding DNA, a protein binding microarray screen was completed using recombinant GST-AP2IV-4 protein (AP2 domain only)(see Material and Methods). The consensus binding sequence from the analysis was determined to be homopolymeric poly(dC):poly(dG); (5’-ACCCCCCT-3’/3’-TGGGGGGA-5’; enrichment score = 0.497) the representative sequence logo shown indicates position weight matrices compiled for each base. An electrophoretic mobility shift assay was completed using 50ng of GST-AP2IV-4 protein and 20 fmol of a 59bp biotinylated DNA probe containing one 6 and one 5 homopolymer poly(dC) stretch (36% total poly(dC) nucleotides). A slowly migrating complex was identified (lane 2, top arrow) that was specifically competed by excess unlabeled poly(dC) competitor DNA (lane 3) but not by excess scrambled DNA fragment containing no more than three consecutive poly(dC) nucleotides (18% poly(dC) nucleotides). (E.) DDAP2IV-4 occupies the native SRS9 promoter in parasite chromatin (see Material and Methods for ChIP-qPCR assay details). Specific DDAP2IV-4 binding was determined in eight specific regions of the SRS9 promoter 5’ to the SRS9 coding region (-1,136 bp flanking including the SRS9 5’-UTR up to the coding ATG). The region of the SRS9 promoter between -584 bp and -192 bp (regions 5–7) showed the highest specific enrichment for DDAP2IV-4 binding. The occurrence of poly(dC) or poly(dG) motifs (5'-3') in the SRS9 promoter are indicated by red or yellow bars, respectively. ChIP-qPCR analysis of DDAP2IV-4 binding to the bradyzoite-specific BAG1 promoter in tachyzoite chromatin was included here as a negative control. A previous study established that the stress-induced ApiAP2 activator, AP2IV-3, specifically binds the BAG1 promoter [28] and is shown here only as a positive control reference. Note that the functionally mapped BAG1 CRE [64] is indicated in the included diagram (yellow diagram). All SRS9 and BAG1 primer sets used in the ChIP-qPCR studies are listed in S3 Dataset.