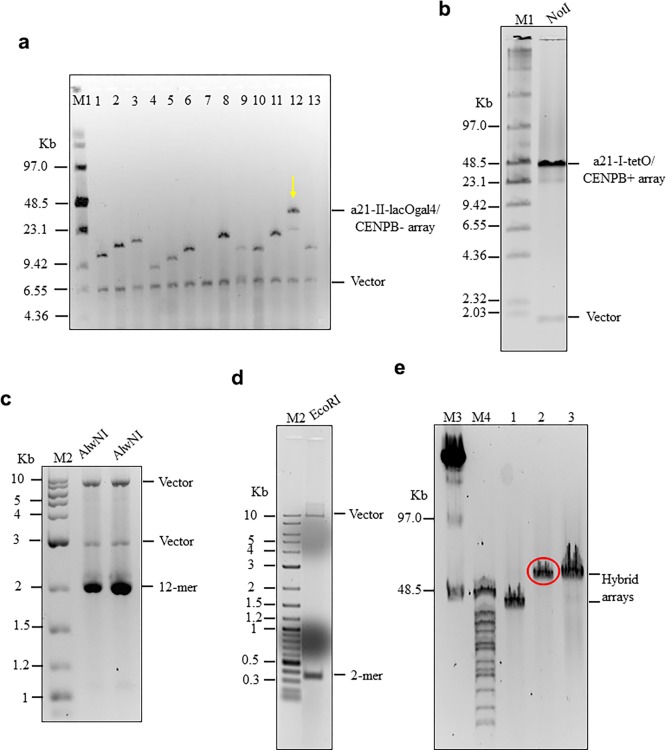
Figure 4.
Hybrid alphoid-DNA array construction. (a) CHEF analysis of 13 BACs with α21-II-lacOgal4 arrays of different size. The BAC DNAs were linearized by AvaII to release a vector part and an array. BAC #12 has an array of ∼40 kb in size. (b) CHEF analysis of the BAC with the α21-I-tetO array of ∼40 kb in size. The BAC DNA was digested by NheI/SpeI to release a vector fragment and the array. (c) Confirmation of the tandem repeat structure of 40 and 60 kb α21-II-lacOgal4 arrays by AlwN1 digestion. CHEF analysis revealed 2,461 bp 12-mer α21-II-lacOgal4 repeat units. (d) Conformation of the tandem repeat structure of a 40 kb α21-I-tetO array by EcoRI digestion. CHEF analysis revealed 343 bp 2-mer 21-I-tetO repeat units. (E) CHEF analysis of the hybrid α21-I-tetO/a21-II-lacOgal4 arrays. (Lane 1) Array consisting of 10 kb of the α21-II-lacOgal4 array and 30 kb of the a21-I-tetO array. (Lane 2) Array consisting of 25 kb α21-II-lacOgal4 array and 30 kb α21-I-tetO array. The array in lane 2 (in red) was chosen for HAC formation. (Lane 3) Array consisting of 15 kb α21-II-lacOgal4 array and 40 kb α21-I-tetO array.