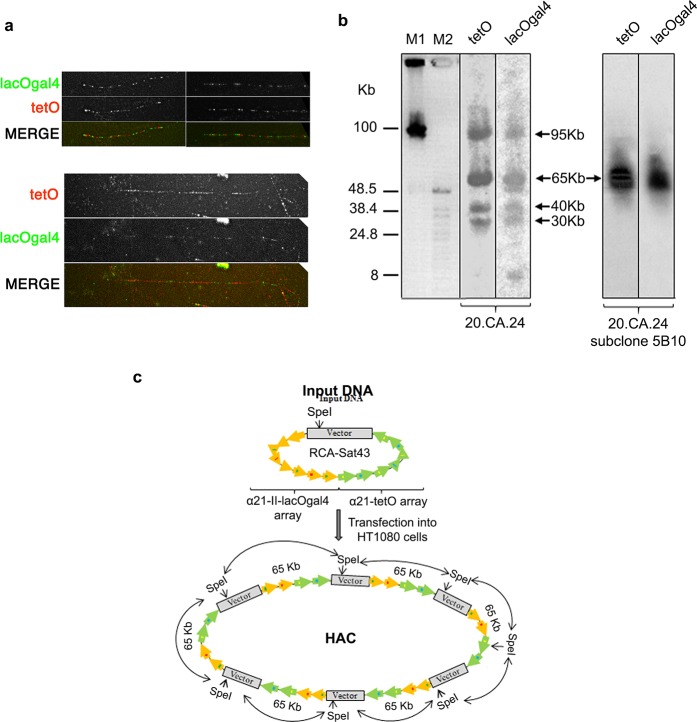
Figure 6.
Structural analysis of the hybrid HAC propagated in human HT1080 cells. (a) Representative fiber-FISH images of clone 20.CA.24 HAC using oligonucleotide probes for tetO (red) and lacO + gal4 sequences (green). Different degrees of fiber stretching are shown (compare upper and lower panels). (b) Genomic DNA possessing the original HAC clone 20.CA.24 (left panel) and its subclone (5B10; right panel) were digested with SpeI endonuclease and separated by CHEF gel electrophoresis (range 10–100 kb). The SpeI recognition site is present once in the RCA-SAT43 vector at position 812 but not in the hybrid array. The transferred membrane was hybridized with radioactively labeled tetO-specific or lacO + gal4-specific probes. The 5B10 subclone has a HAC with a remarkably conserved array. Arrows indicate fragments of 95, 65, 40, and 30 kb in size that are specific to both probes. (c) Diagram illustrating multimerization of input DNA during de novo HAC formation in human HT1080 cells. Input DNA consists of 65 kb hybrid array and 10,209 bp RCA-Sat43 vector sequence.