Fig 3. Mitoribosome mtLSU protein stability correlates with structural modules and defines an assembly pathway. See also Figure S3.
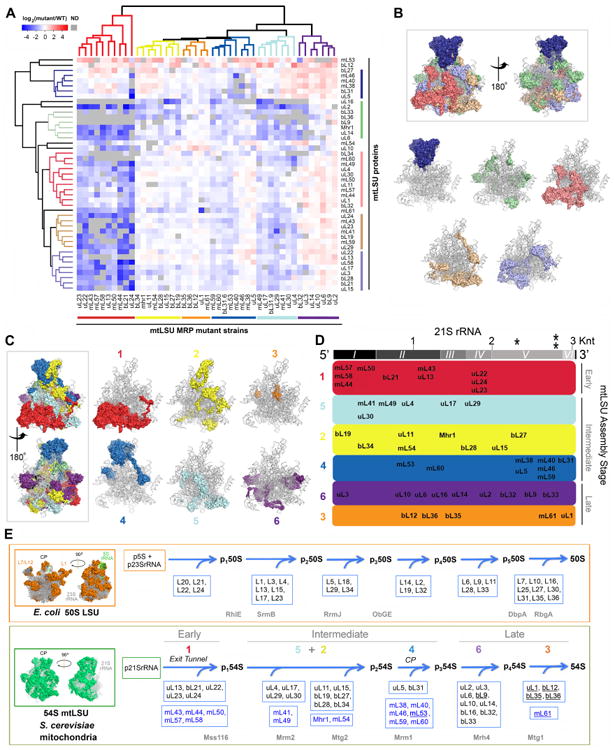
A. Following analysis by mass spectrometry of the mtLSU intermediates that accumulate in the mtLSU mutant strains (fractions indicated in Fig. 2C), the data obtained on the abundance of mtLSU subunits accumulated in mtLSU subassemblies was used for cluster analysis of MRP mutant strains (see STAR Methods). The several clusters identified are color-coded. The results presented are representative of two independent biological replicates. ND, not detected.
B. Clusters of mtLSU proteins defined in panel A (vertical clustering), mapped to the mtLSU structure (PDB-3J6B (Amunts et al., 2014)) with 21S rRNA shown in gray. The top portion presents two views of the mtLSU with all the clusters color-coded, and the lower portion presents the location of individual clusters on the 21S rRNA backbone.
C. mtLSU MRP mutant strains clusters defined in panel A (horizontal clustering) mapped to the mtLSU structure with 21S rRNA shown in gray. The left portion presents two views of the mtLSU with all the clusters color-coded, and the right portion presents the location of individual clusters (specified as 1 to 6) on the 21S rRNA backbone.
D. mtLSU assembly map following the modules defined in panel C. The proteins are distributed along the length of the 21S rRNA (grey bar on top), based on the protein-RNA interactions defined in the mtLSU structure (Amunts et al., 2014). The secondary structure domains of 21S rRNA are indicated as I to VI. * and ** indicate the two methylation sites in the 21S rRNA (Gm2270 and Um2791). Knt, kilonucleotides. The clusters are ordered following their incorporation during early, intermediate or late stages of mtLSU assembly.
E. Comparison of the mtLSU assembly map proposed here with the known assembly map of bacterial (E. coli) LSU (Chen and Williamson, 2013). On the left, the structure cartoons of the bacterial and yeast mitochondria LSU are presented. In the proposed yeast mtLSU assembly pathway, in each cluster, proteins conserved in bacteria are boxed and marked in black, and mitochondria-specific proteins are boxed and labeled in blue. Names of non-essential proteins are underlined. The clusters proposed to assemble during intermediate stages are separated into 5+2 and 4 (corresponding to the central protuberance (CP)), given the reported independent existence of a stable CP module (Box et al., 2017). At the bottom of the yeast 54S mtLSU assembly pathway are listed in grey the 6 assembly factors so far described. Proposed bacterial orthologs and paralogs of these assembly factors are listed in grey at the bottom of the 50S E. coli LSU assembly pathway.
