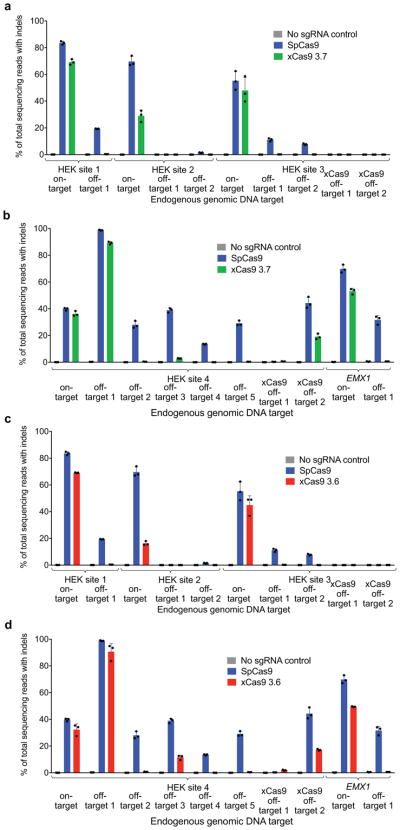
Extended Data Figure 11. Validation by high-throughput sequencing of GUIDE-seq results.
The most frequent off-target sites identified by GUIDE-seq were verified by HTS of genomic DNA following treatment of HEK293T cells with SpCas9 or xCas9 3.7 (a, b), or following treatment with SpCas9 or xCas9 3.6 (c, d). New sites with non-NGG PAMs that were identified as xCas9 off-target sites were also analyzed in (a–d). Values and error bars reflect the mean and s.d. of n=3 biologically independent samples. Target sequences are in Supplementary Table 16.