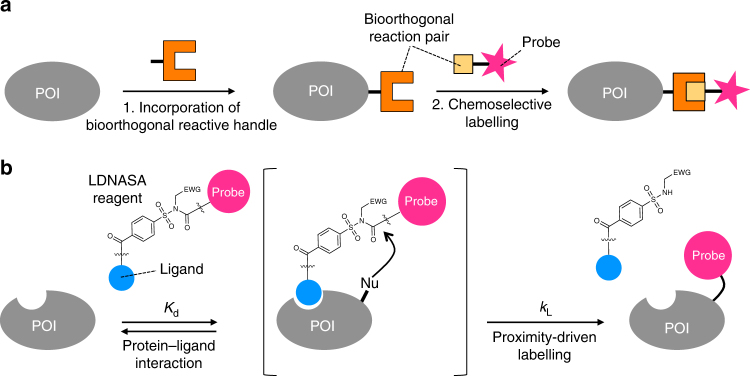
Fig. 1.
Two distinct approaches for bioorthogonal protein labelling. a Schematic illustration of protein labelling through bioorthogonal chemistry. The first step involves the genetic incorporation of a bioorthogonal reactive handle into a protein of interest (POI) in cells. In a second step, the reactive handle chemoselectively reacts with a designed synthetic probe. b Schematic illustration of the basic principle of ligand-directed N-acyl-N-alkyl sulfonamide (LDNASA) chemistry. The reagent binds to a POI through a specific protein–ligand interaction, driving a chemical reaction between the reactive group and a natural nucleophilic amino acid located on the protein surface, through the proximity effect. EWG, electron-withdrawing group; Kd, dissociation constant; kL, first-order rate constant for the labelling process; Nu, nucleophilic amino acid