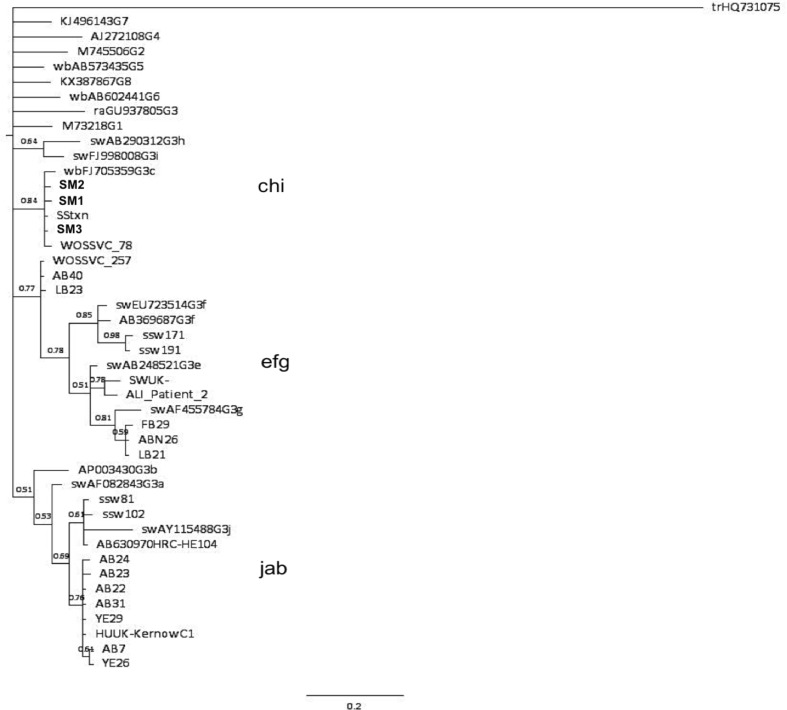
Fig. 1.
Phylogenetic analysis of 145-nt open reading frame 2 (ORF2) fragments (relative to Burma strain M73218 positions 6390-6534) from Scottish commercial mussels (in bold). Numbers beside nodes indicate posterior probabilities. Reference sequences for all HEV genotypes and subtypes are as follows—genotype 1, M73218; 2, M74506; 3a, AF082843; 3b, AB291955; 3c, FJ705359; 3e, AB248521; 3f, AB369687; 3f, EU723514; 3g, AF455784; 3h, AB290312; 3i, FJ998008; 3j, AY115488; G4, AJ272108; G3 rabbit, GU937805; G5 wild boar, AB5734435; G6 wild boar, AB602441; G7 camel, KJ496143; Cutthroat trout strain Heenan88, HQ389543 (Smith et al. 2016). Additional sequences included in analysis—huUK—human HEV strain C1 Kernow, camel G8, KX8387867. Abbreviations included in tree are as follows—sw swine, ssw Scottish swine, wb wild boar, ra rabbit. ORF2 sequences obtained from Scottish wild mussels included in the phylogenetic analysis are named based on their source of origin and are as follows: AB Ardrossan beach, ABN Aberdeen, YE Ythan estuary, FB Ferrybridge, LB Lunderston Bay (Crossan et al. 2012). ORF2 sequences from Scottish patient sera have also been included in the analysis and are labelled as follows; ALI_patient_2; WOSSVC_78; WOSSVC_257; SStxn (accession number KT159771.1). HEV subtype clusters are indicated by the groupings of letters. Phylogenetic tree analysis was performed using MrBayes version 3.2.1 and the phylogenetic tree generated using Figtree version 1.4.2