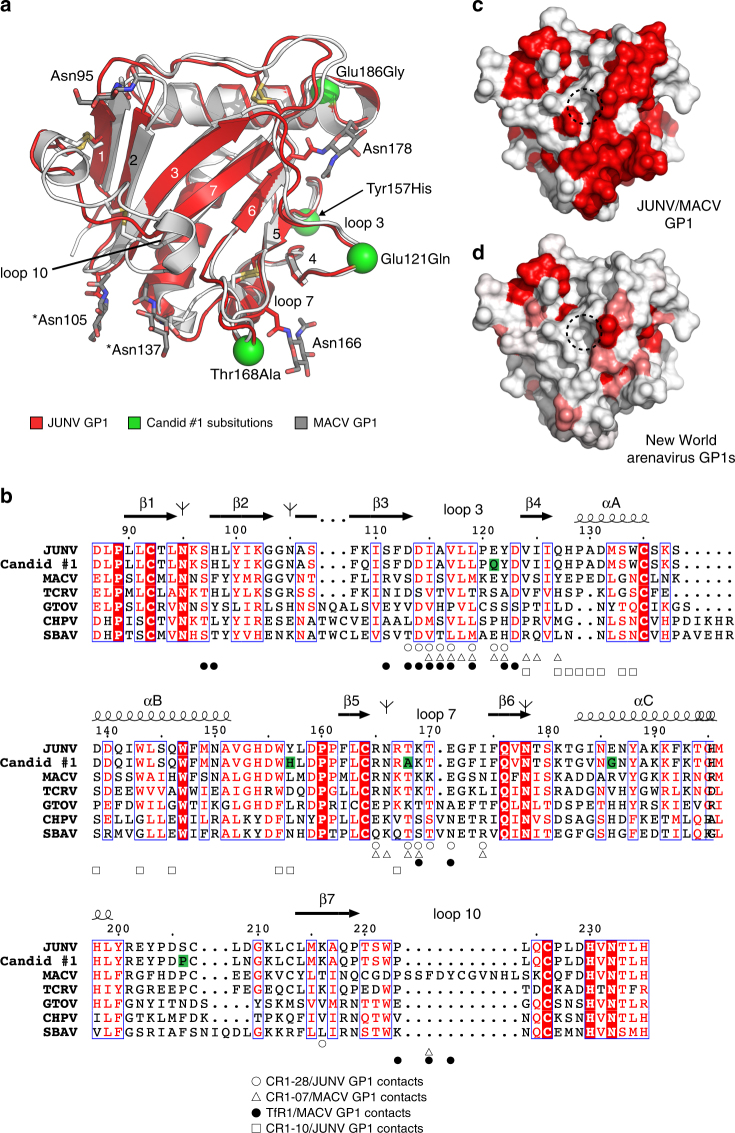
Fig. 5.
Sequence diversity in New World arenavirus GP1s. a Overlay of ribbon diagrams of JUNV GP1 (red, PDB: 5EN214) and MACV GP1 (gray, PDB: 3KAS13). N-linked glycans are shown as sticks. Asterisks indicate N-linked glycans that differ in position between JUNV and MACV. Green spheres are sites of substitutions between the JUNV MC2 strain and Candid #1, the live attenuated vaccine strain. Loop 10 is a disulfide-linked insert that is unique to MACV. Disulfide bridges are shown as yellow sticks. b Sequence alignment of New World arenavirus GP1s. Antibody and TfR1 contacts are indicated. Substitutions in Candid #1 (with respect to JUNV MC2) are highlighted in green. Tree diagrams indicate predicted sites of N-linked glycosylation for JUNV GP1. The figure was generated using ESPrit364 and modified. GenBank accession numbers used for sequences used for the alignment are JUNV (MC2, D10072), JUNV (Candid #1, ACY70854.1), MACV (Carvallo, NC_005078), GTOV (INH-95551, NC_005077), CHPV (810419, YP_001816782.1), and SBAV (SPH114202, NC_006317). The sequence of TCRV shown in the alignment is that of the pseudotype used in this study and other studies,14, 30 which has a deletion spanning amino acid residues 121–132 and three substitutions (I134A, G418S and E458R) compared to GenBank NC_004293. c Surface representation of JUNV GP1 (PDB: 5EN2)14 showing conservation based on the alignment with MACV GP1 shown in b generated with ESPrit3.64 Dashed circle indicates the location of the Tyr211TfR1 pocket. d Surface representation of JUNV GP1 (PDB: 5EN2)14 showing sequence conservation with MACV, GTOV, CHPV, SBAV as shown in b. Using the algorithm available in ESPrit3,64 residues with a similarity global score lower than 0.7 are in white, and residues with a similarity global score in the range 0.7–1.0 are color-ramped in shades of red (with dark red showing the greatest conservation)