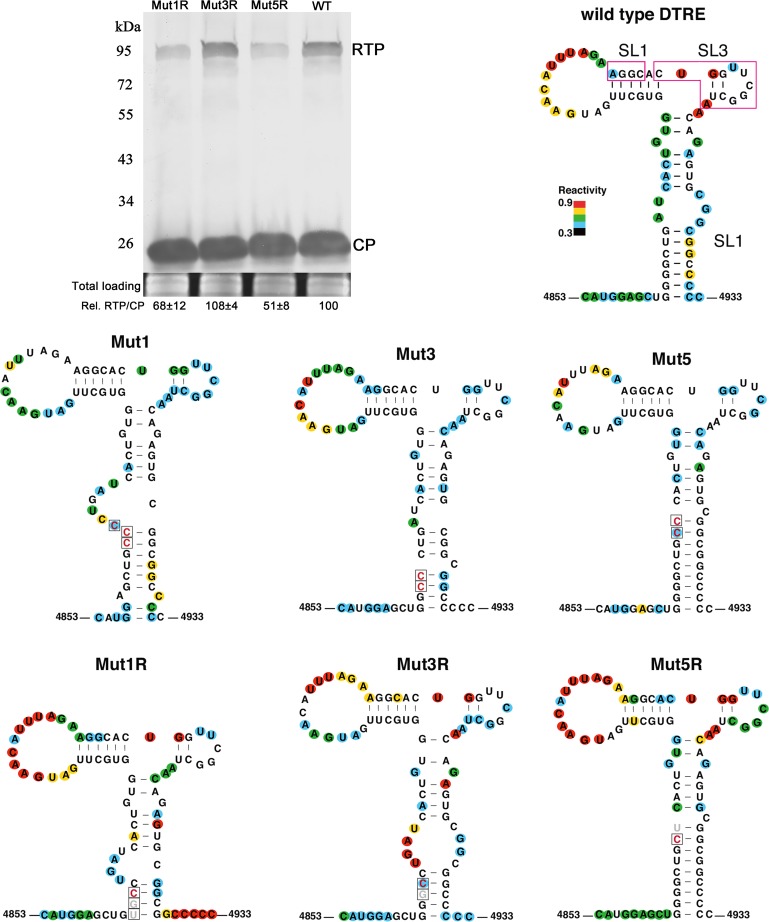
FIG 5.
Secondary structures of the distal translation elements from wild-type virus (WT) and from stem-loop mutants Mut1, Mut3, and Mut5. The blot shows RTP translation by Mut1R, Mut3R, and Mut5R in agroinfiltrated N. benthamiana tissues at 3 dpi. Relative readthrough (Rel. RTP/CP) was calculated as the RTP/CP ratio, with that for wild-type PLRV set as 100%. Values represent the means (± standard error) determined from three independent experiments. Secondary structures of wild-type, mutant, and second-site mutant (“revertant”) DRTEs that were recovered from plants that were symptomatic at 7 to 8 wpi are also shown. Each RNA was subjected to SHAPE probing, from which structure was predicted using SAFA and MFOLD software (see Materials and Methods). The degree of chemical modification (“single-strandedness”) of each base is indicated by color-coded circles, with red indicating the most modified and blue the least. Uncolored bases showed no modification. Mutations are boxed, with constructed mutations in red and revertants in gray. Magenta boxes on wild-type RNA indicate bases predicted to base pair to the C-rich region adjacent to the leaky stop codon (Fig. 7).