Figure 5.
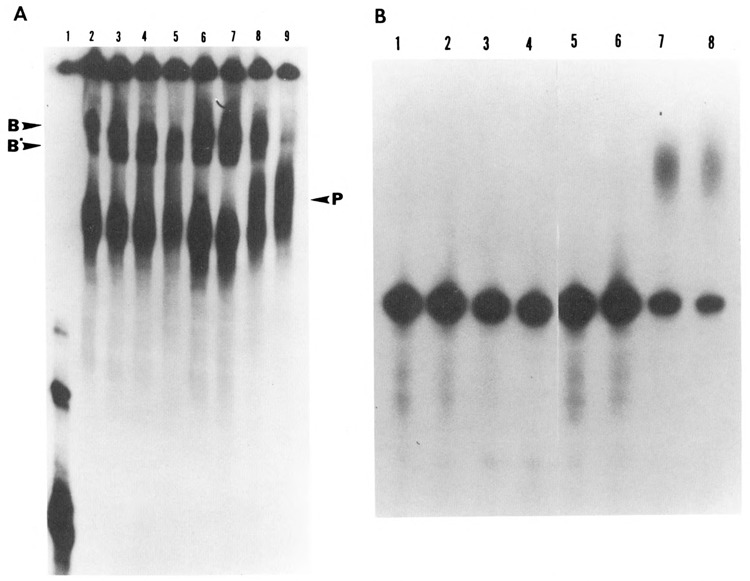
Effect of dephosphorylation on the formation of RNP complexes under polyadenylation conditions. HeLa nuclear extract was dephosphorylated with alkaline phosphatase (0.04 U/μg). Control samples consisted of extract incubated with 10 mM Hepes pH 7.6 in a volume equal to that of treated samples. Polyadenylation reactions were carried out for varying time intervals ranging from 0 to 90 minutes using [32P]-labeled SV40 pre-mRNA substrate. Half of the reaction (12.5 μl) was terminated by the addition of heparin to a final concentration of 5 mg/ml and incubated for an additional 10 minutes at 30°C. These samples were analyzed on a 4% non-denaturing polyacrylamide gel at 4°C (A). The remaining samples were deproteinized and analyzed on an 8% acrylamide, 8.3 M Urea gel (B). A. Lanes 2 to 5 correspond to the dephosphorylated samples incubated for 0, 5, 30, and 90 minutes respectively, while lanes 6 to 9 correspond to the untreated sample incubated for the same time periods. Electrophoresis on the native gel showed that in the primary stages of the RNP complex formation, where cleavage takes place, the dephosphorylated and non-dephosphorylated samples formed comparable complexes with pre-mRNA (compare lanes 2 and 3 with lanes 6 and 7). However, as the reaction progressed from cleavage to polyadenylation, the treated samples were unable to form a final polyadenylated RNP complex (compare lanes 4 and 5 with lanes 8 and 9). The arrow on the left corresponds to pre-mRNA used as the substrate. B. Electrophoresis of the remaining half of the samples under denaturing conditions showed the state of the RNA corresponding to the complexes formed in A. The emergence of the polyadenylated RNA correlated with the formation of the polyadenylation-specific complexes (P); compare B, lanes 7 and 8 with A, lanes 8 and 9. The sample corresponding to A, lane 1 was not processed under denaturing conditions, as it represented only the labeled pre-mRNA probe.
