Figure 4.
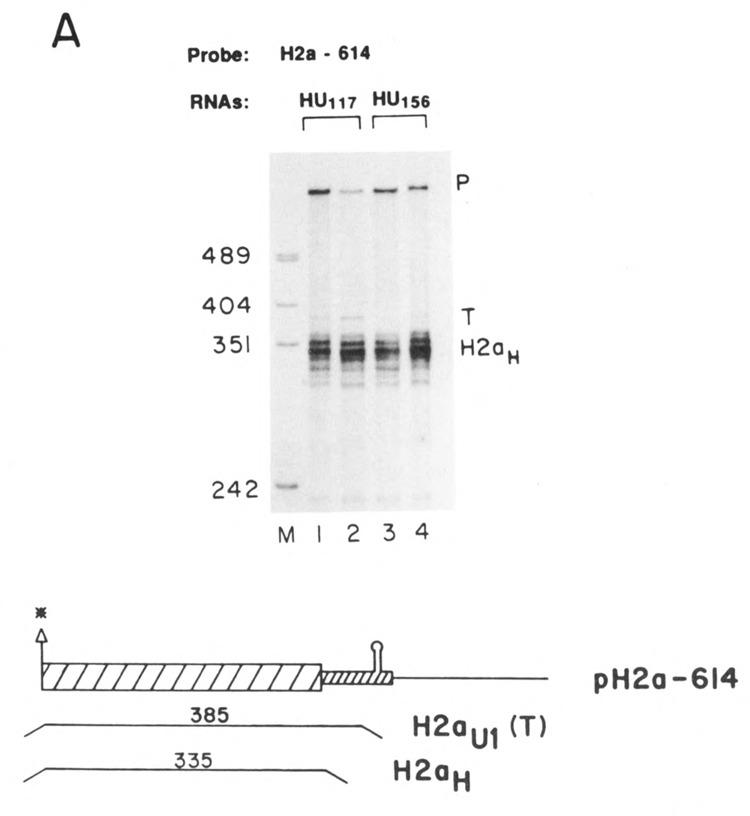
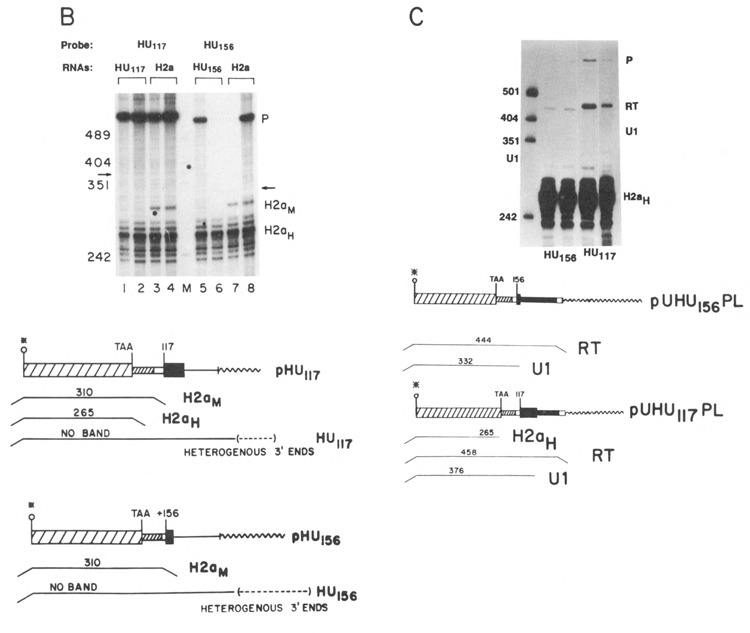
Expression of the HU genes. A. Duplicate preparations of RNA from cells transfected with the HU117 genes (lanes 1 and 2) or the HU156 genes (lanes 3 and 4) were analyzed by S1 nuclease mapping using the H2a-614 gene labeled at the 3′ end of the Aval site as a probe. This probe will map all transcripts from the HU genes which extend more than 60 nucleotides past the 3′ end of the histone coding region as a single protected fragment (labeled T). The other protected fragment (H2aH) is due to protection of the endogenous hamster H2a mRNAs; B. Duplicate preparations of RNA (5 μg) from cells transfected with the HU117 (lanes 1 and 2) or the HU156 genes (lanes 5 and 6) were analyzed by S1 nuclease mapping using the HU117 (lanes 1–4) or HU156 (lanes 5–8) genes labeled at the 3′ end of the Narl site at codon 43 as probes. RNA from cells transfected with the H2a-614 gene was analyzed in lanes 3, 4, 7, and 8. The arrows indicate the expected positions of transcripts ending at the U1 3′ end (332 and 376 nucleotides respectively). Band P is undigested probe. C. RNAs from cells transfected with the HU156; or the HU117 were analyzed by SI nuclease mapping using the probes UHU117PL or UHUnvPL respectively. The RNAs used are indicated beneath the lanes. These probes have a polylinker insert either 112 or 82 nucleotides after the U1 3′ end, which allows the mapping of longer heterogenous read-through transcripts from these genes as a single protected fragment (labeled RT). The position of the protected fragment which would correspond to the U1 3′ end is indicated. The protected fragment labeled H2aH corresponds to protection of the endogenous hamster H2a mRNAs. Lane M is marker pUC18 digested with HpaII. P is the position of the undigested probe.
