FIG 4.
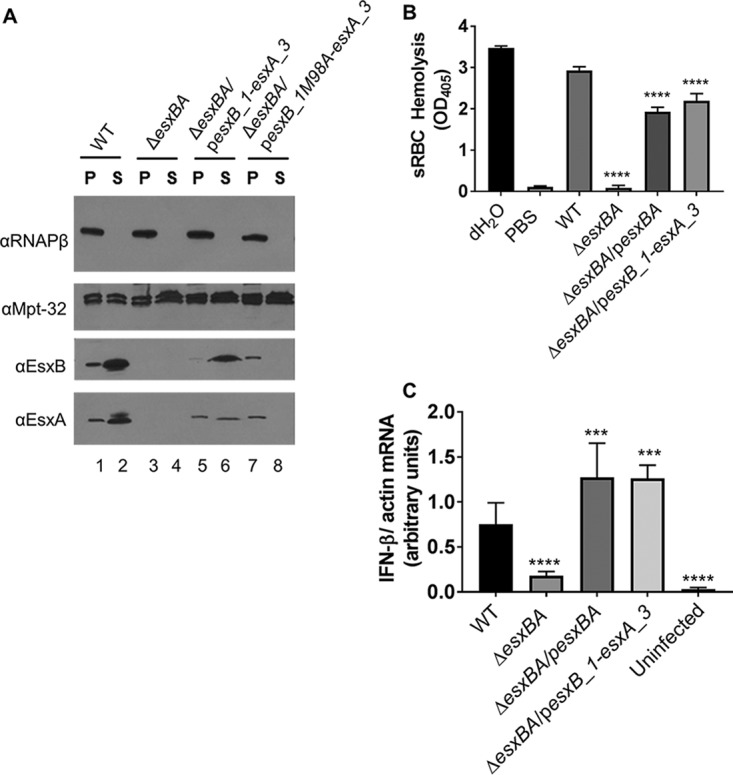
EsxB_1 and EsxA_1 can by secreted by Esx-1 and are functionally redundant. (A) ESX-1 secretion assay. Controls for the secretion assay are the same as those described in the legend to Fig. 2. In this assay, the EsxB and EsxA antibodies are recognizing EsxB_1 and EsxA_1/EsxA_3 for lanes 5 to 8. Data are representative of at least three biological replicates. (B) sRBC hemolysis assay. The data represent at least four biological replicates, each with three technical replicates. The data were averaged, and the propagated error was calculated from the standard deviation from each replicate. The error bars represent the propagated error. Significance was calculated using an ordinary one-way ANOVA (P < 0.0001) followed by Dunnett's multiple comparison for results compared to those with the WT strain (****, P < 0.0001). (C) qRT-PCR analysis measuring IFN-β expression normalized to actin expression at 24 h postinfection with M. marinum. The data are the average of four biological replicates, each with two technical replicates. The error bars represent the propagated error (n = 8 total). Statistical significance was determined using an ordinary one-way ANOVA (P < 0.0001) followed by Dunnett's multiple-comparison test comparing results with infection by the WT strain. (****, P = 0.0001; ***, P = 0.0003 and 0.0002 for pesxBA and pesxB_1-esxA_3, respectively).
