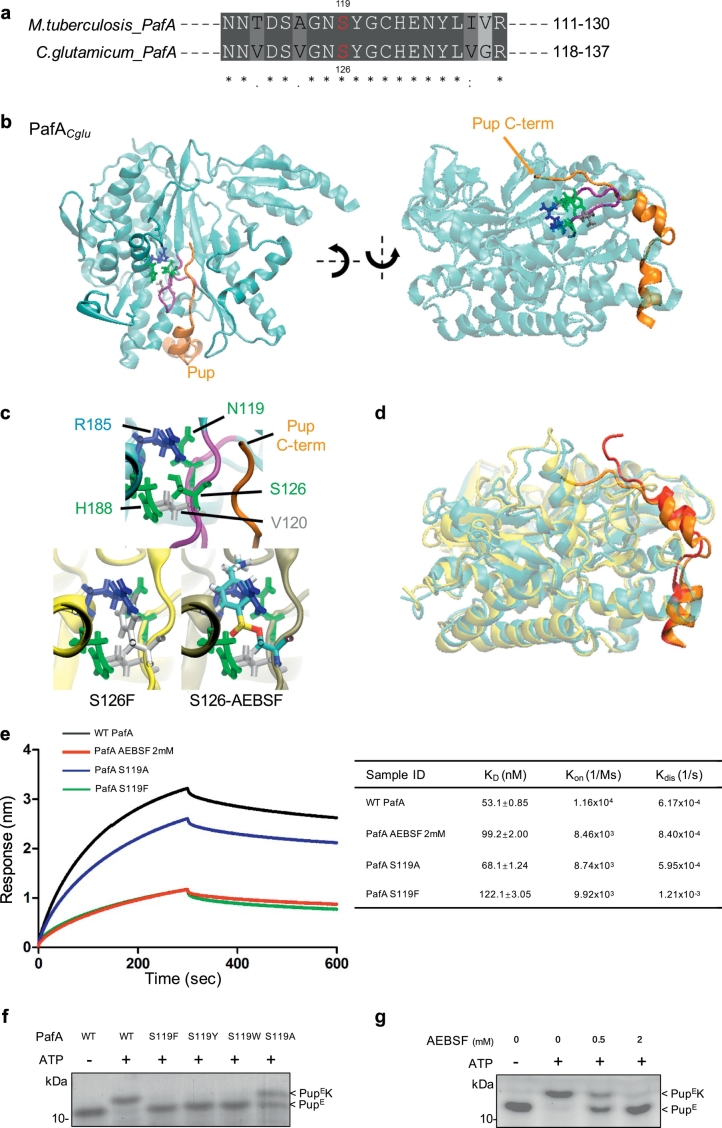
Fig. 4.
Structural basis for the involvement of S119 in Mtb PafA activity.
(a) Alignment of residues 111–130 of Mtb PafA to that of PafA from C. glutamicum. Sequences were compiled from the National Center for Biotechnology Information server and aligned by means of ClustalW.
(b) Crystal structure of the C. glutamicum PafA/Pup complex. Pup (orange) wraps around PafA (cyan) via two small α-helices and a short C-terminal tail. Binding of Pup to substrate proteins occurs via its C-terminus. Also shown in stick representation are the S126 pocket residues and the S126-loop (purple) that define an extended interface of the Pup-binding groove of PafA.
(c) Close-up view of the S126-pocket of the WT, S126F and S126-AEBSF initial structures. Simply changing the S126 side chain to phenylalanine or attaching AEBSF results in significant steric overlap with other pocket resides. Thus, these modifications must disrupt the organization of this pocket. Phenylalanine is colored white and the carbon, oxygen, sulfur, nitrogen, and hydrogen atoms in the AEBSF-linked residue are colored cyan, red, yellow, blue, and white, respectively.
(d) Snap-shot of the equilibrium MD simulations of the WT and S126F PafACglu/Pup complexes, with the PafA proteins overlapped. The interaction of PafA with Pup is significantly weakened in S126F as a result of the disruption of the S126-pocket compared with the WT. This weakened interaction occurs at both the Pup C-terminal tail, which directly contacts the S126-loop, and its α-helices, which are distant from the S126-loop.
(e) BLI data for the binding of PupE to PafA variants or PafA pre-incubated with 2 mM AEBSF and their interaction kinetics. PupE was immobilized on streptavidin-coated biosensors and exposed to binding partners in buffer. Binding was measured by coincident changes in the interference pattern. The KD (nM), Kon (1/Ms) and Kdis (1/s) are shown in the table to the right.
(f) Pupylation reactions included free lysine (80 mM), PupE (10 μM) and PafA variants (0.5 μM each) and were incubated at 25 °C for 15 min with 5 mM ATP in pupylation buffer. Samples were analyzed by SDS-PAGE, followed by CBB staining.
(g) As in Fig. 4f, except that PafA (0.5 μM) pre-incubated with AEBSF at 25 °C for 0.5 h was used instead of PafA variants.