Fig. 5.
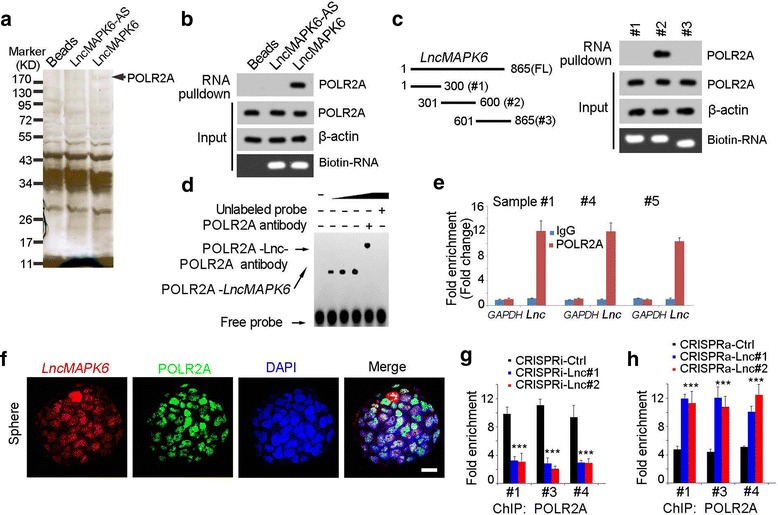
LncMAPK6 interacted with RNA polymerase II. (a) LncMAPK6 was transcribed in vitro for RNA pulldown, and the denoted band in lncMAPK6 sample was identified as POLR2A. (b) The binding of lncMAPK6 and POLR2A was examined by Western blot. (c) LncMAPK6 truncates were generated (left panels), and incubated with sphere lysates. The interaction between lncMAPK6 truncates and POLR2A was confirmed (right panels). (d) RNA EMSA was performed for the combination between lncMAPK6 and POLR2A. The second truncate of lncMAPK6 was used for RNA EMSA. (e) Oncospheres derived from clinical samples were used for RNA immunoprecipitation (RIP) assay, and enrichment of lncMAPK6 were detected with realtime PCR. GAPDH was a control. (f) The co-localization of lncMAPK6 and POLR2A was confirmed by double FISH assay. Scale bars, 10 μm. (g, h) LncMAPK6 depleted (g) and overexpressed (h) cells were crushed for POLR2A ChIP, followed by detection for MAPK6 promoter enrichment with realtime PCR
