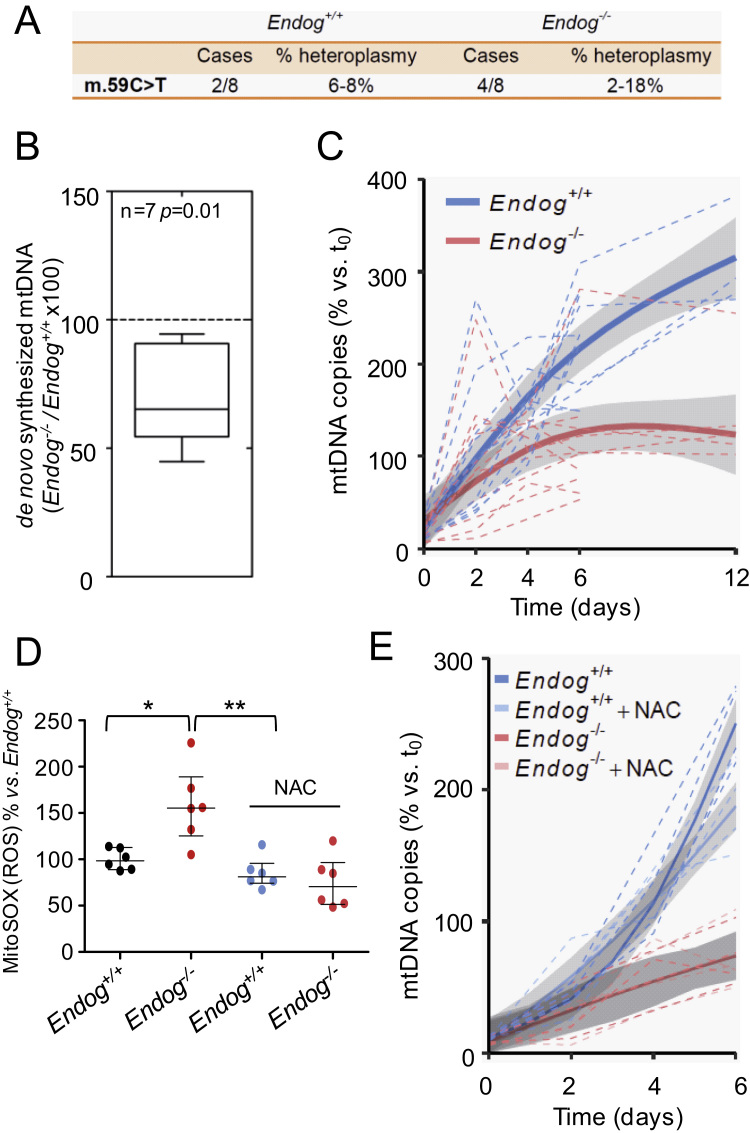
Fig. 3.
EndoG deficiency has no effect on mtDNA stability but hinders mtDNA synthesis independently of ROS accumulation. (A) Sequencing of mitochondrial DNA extracted from hearts of Endog+/+ and Endog-/- neonatal mice did not show any nucleotide variant associated to the Endog genotype. A single mutation (59 C>T) was detected in heteroplasmy (% frequency vs. reference sequence) within the Phe tRNA (MT-TF) gene in 2 Endog+/+ and 4 Endog-/- mice; N=8 per genotype, ns: not statistically significant differences. (B) Incorporation of dTTP-3H in mtDNA was quantified simultaneously in isolated Endog+/+ and Endog-/- cardiac mitochondria preparations; N=7. Non-parametric sign test for paired observations indicated significant differences due to genotype, P=0.01. (C) MtDNA of MEF from Endog+/+ and Endog-/- mice was depleted with EtBr (25 ng/ml) in order to reach minimum similar levels in both genotypes. Removal of the drug from the culture medium (day 0) allowed recovery of mtDNA content and the mtDNA copy number was determined in Endog+/+ and Endog-/- MEF at the indicated time intervals. The relative mtDNA copy number was quantitated and analyzed as described in the Materials and Methods section. N=5 per genotype. (D) Mitochondrial ROS abundance was quantified using the MitoSOXTM mitochondrial superoxide indicator in Endog+/+ and Endog-/- fibroblasts cultured in the absence or presence of ROS scavenger NAC (0.2 mM). Due to important inter-experiment basal signal oscillations, data were normalized divinding by the Endog+/+ mean before statistical treatment. The Kruskall-Wallis test (P=0.03) was followed by the Dunn's test for pair wise comparisons; *, P<0.05 vs. Endog+/+; **, P<0.01 vs. Endog+/++NAC. Medians ± interquartile ranges are shown. N=6. (E) MtDNA copy number recovery was analyzed in the presence of ROS scavenger NAC. After removal of BrEt (day 0), Endog+/+ and Endog-/- MEF were divided into two groups during one week recovery period. In both genotypes, one group was cultured in a medium with NAC (0.2 mM) and the other group served as control without NAC. At the indicated time intervals, the relative mtDNA copy number was determined in cells from each group as in Fig. 3.C. N=5 per genotype.